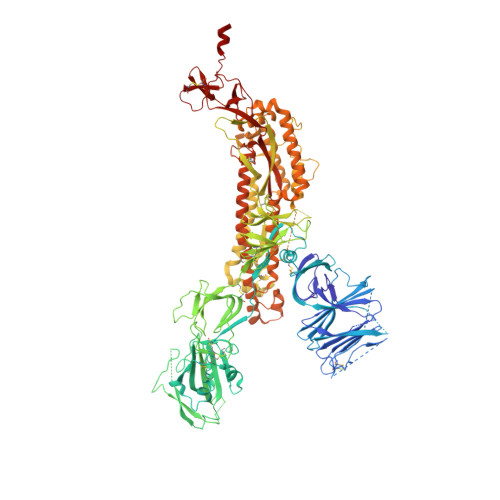
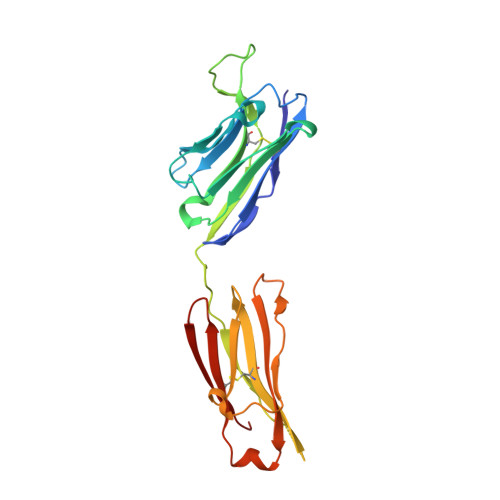
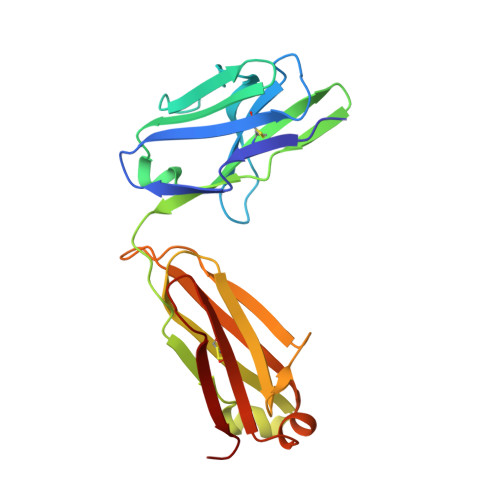
Fab-dimerized glycan-reactive antibodies are a structural category of natural antibodies.
Williams, W.B., Meyerhoff, R.R., Edwards, R.J., Li, H., Manne, K., Nicely, N.I., Henderson, R., Zhou, Y., Janowska, K., Mansouri, K., Gobeil, S., Evangelous, T., Hora, B., Berry, M., Abuahmad, A.Y., Sprenz, J., Deyton, M., Stalls, V., Kopp, M., Hsu, A.L., Borgnia, M.J., Stewart-Jones, G.B.E., Lee, M.S., Bronkema, N., Moody, M.A., Wiehe, K., Bradley, T., Alam, S.M., Parks, R.J., Foulger, A., Oguin, T., Sempowski, G.D., Bonsignori, M., LaBranche, C.C., Montefiori, D.C., Seaman, M., Santra, S., Perfect, J., Francica, J.R., Lynn, G.M., Aussedat, B., Walkowicz, W.E., Laga, R., Kelsoe, G., Saunders, K.O., Fera, D., Kwong, P.D., Seder, R.A., Bartesaghi, A., Shaw, G.M., Acharya, P., Haynes, B.F.(2021) Cell 184: 2955
- PubMed: 34019795
- DOI: https://doi.org/10.1016/j.cell.2021.04.042
- Primary Citation of Related Structures:
6VTU, 6XRJ, 7L02, 7L06, 7L09, 7L6M, 7L6O, 7LU9, 7LUA - PubMed Abstract:
Natural antibodies (Abs) can target host glycans on the surface of pathogens. We studied the evolution of glycan-reactive B cells of rhesus macaques and humans using glycosylated HIV-1 envelope (Env) as a model antigen. 2G12 is a broadly neutralizing Ab (bnAb) that targets a conserved glycan patch on Env of geographically diverse HIV-1 strains using a unique heavy-chain (V H ) domain-swapped architecture that results in fragment antigen-binding (Fab) dimerization. Here, we describe HIV-1 Env Fab-dimerized glycan (FDG)-reactive bnAbs without V H -swapped domains from simian-human immunodeficiency virus (SHIV)-infected macaques. FDG Abs also recognized cell-surface glycans on diverse pathogens, including yeast and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spike. FDG precursors were expanded by glycan-bearing immunogens in macaques and were abundant in HIV-1-naive humans. Moreover, FDG precursors were predominately mutated IgM + IgD + CD27 + , thus suggesting that they originated from a pool of antigen-experienced IgM + or marginal zone B cells.
Organizational Affiliation:
Duke Human Vaccine Institute, Durham, NC 27710, USA; Department of Medicine, Duke University, Durham, NC 27710, USA. Electronic address: wilton.williams@duke.edu.