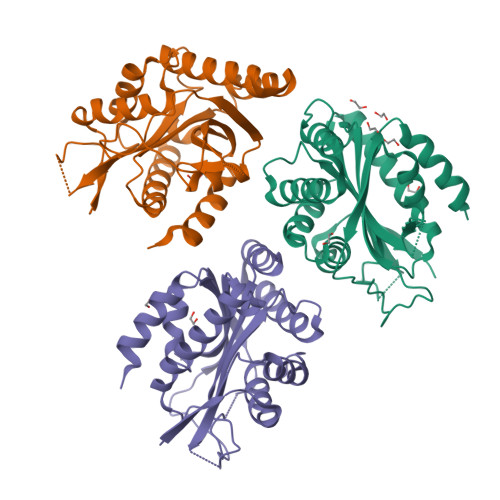
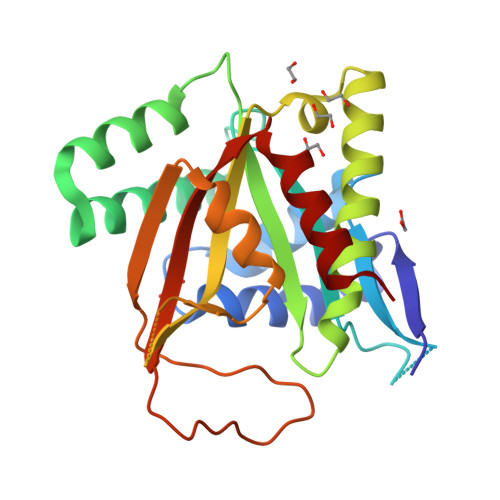
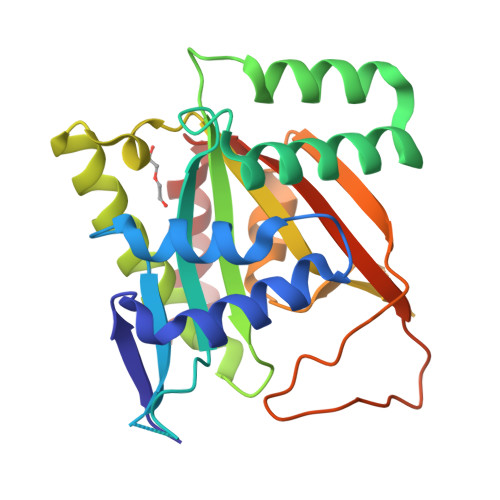
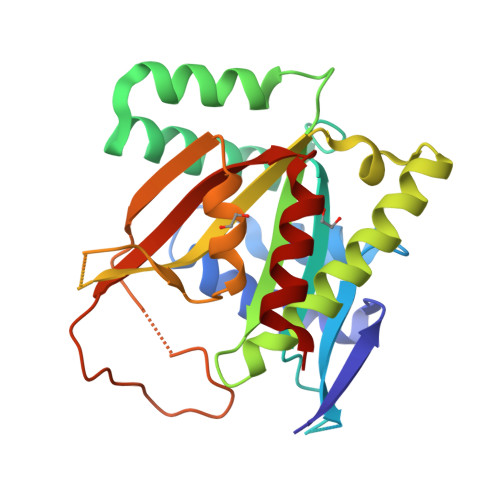
Structure of human BCCIP and implications for binding and modification of partner proteins.
Choi, W.S., Liu, B., Shen, Z., Yang, W.(2021) Protein Sci 30: 693-699
- PubMed: 33452718
- DOI: https://doi.org/10.1002/pro.4026
- Primary Citation of Related Structures:
7KYQ, 7KYS - PubMed Abstract:
BCCIP was isolated based on its interactions with tumor suppressors BRCA2 and p21. Knockdown or knockout of BCCIP causes embryonic lethality in mice. BCCIP deficient cells exhibit impaired cell proliferation and chromosome instability. BCCIP also plays a key role in biogenesis of ribosome 60S subunits. BCCIP is conserved from yeast to humans, but it has no discernible sequence similarity to proteins of known structures. Here we report two crystal structures of an N-terminal truncated human BCCIPβ, consisting of residues 61-314. Structurally BCCIP is similar to GCN5-related acetyltransferases (GNATs) but contains different sequence motifs. Moreover, both acetyl-CoA and substrate-binding grooves are altered in BCCIP. A large 19-residue flap over the putative CoA binding site adopts either an open or closed conformation in BCCIP. The substrate binding groove is significantly reduced in size and is positively charged despite the acidic isoelectric point of BCCIP. BCCIP has potential binding sites for partner proteins and may have enzymatic activity.
Organizational Affiliation:
Laboratory of Molecular Biology, NIDDK, National Institutes of Health, Bethesda, Maryland, USA.