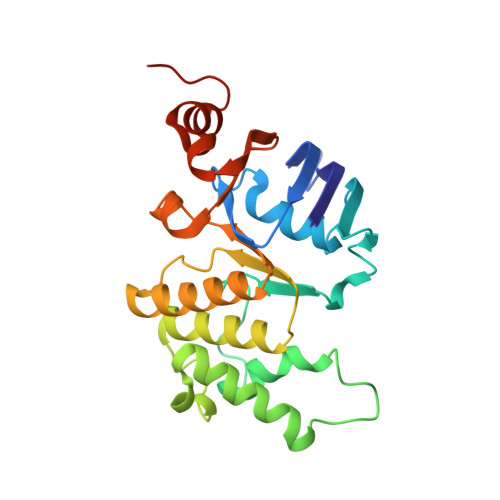
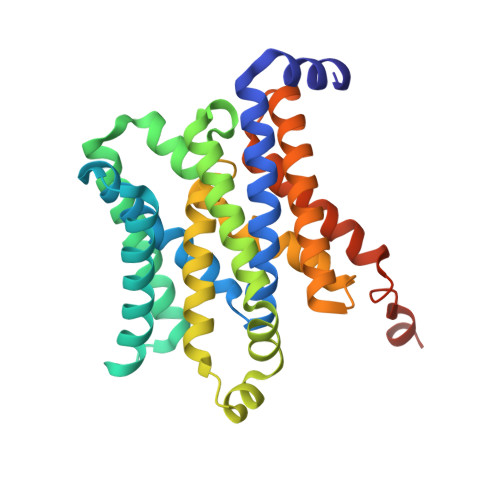
The structural basis of bacterial manganese import.
Neville, S.L., Sjohamn, J., Watts, J.A., MacDermott-Opeskin, H., Fairweather, S.J., Ganio, K., Carey Hulyer, A., McGrath, A.P., Hayes, A.J., Malcolm, T.R., Davies, M.R., Nomura, N., Iwata, S., O'Mara, M.L., Maher, M.J., McDevitt, C.A.(2021) Sci Adv 7
- PubMed: 34362732
- DOI: https://doi.org/10.1126/sciadv.abg3980
- Primary Citation of Related Structures:
7KYO, 7KYP - PubMed Abstract:
Metal ions are essential for all forms of life. In prokaryotes, ATP-binding cassette (ABC) permeases serve as the primary import pathway for many micronutrients including the first-row transition metal manganese. However, the structural features of ionic metal transporting ABC permeases have remained undefined. Here, we present the crystal structure of the manganese transporter PsaBC from Streptococcus pneumoniae in an open-inward conformation. The type II transporter has a tightly closed transmembrane channel due to "extracellular gating" residues that prevent water permeation or ion reflux. Below these residues, the channel contains a hitherto unreported metal coordination site, which is essential for manganese translocation. Mutagenesis of the extracellular gate perturbs manganese uptake, while coordination site mutagenesis abolishes import. These structural features are highly conserved in metal-specific ABC transporters and are represented throughout the kingdoms of life. Collectively, our results define the structure of PsaBC and reveal the features required for divalent cation transport.
Organizational Affiliation:
Department of Microbiology and Immunology, The Peter Doherty Institute for Infection and Immunity, The University of Melbourne, Melbourne, Victoria, Australia.