Novel Bisubstrate Inhibitors for Protein N-Terminal Acetyltransferase D.
Deng, Y., Deng, S., Ho, Y.H., Gardner, S.M., Huang, Z., Marmorstein, R., Huang, R.(2021) J Med Chem 64: 8263-8271
- PubMed: 34110812
- DOI: https://doi.org/10.1021/acs.jmedchem.1c00141
- Primary Citation of Related Structures:
7KD7, 7KPU - PubMed Abstract:
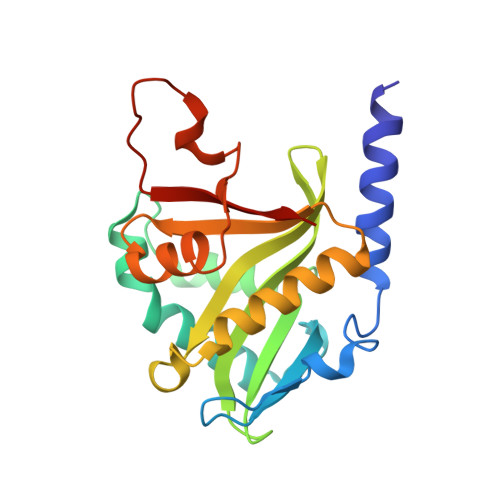
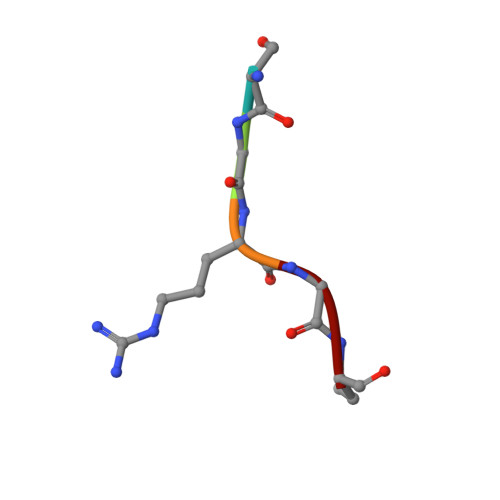
Protein N-terminal acetyltransferase D (NatD, NAA40) that specifically acetylates the alpha-N-terminus of histone H4 and H2A has been implicated in various diseases, but no inhibitor has been reported for this important enzyme. Based on the acetyl transfer mechanism of NatD, we designed and prepared a series of highly potent NatD bisubstrate inhibitors by covalently linking coenzyme A to different peptide substrates via an acetyl or propionyl spacer. The most potent bisubstrate inhibitor displayed an apparent K i value of 1.0 nM. Biochemical studies indicated that bisubstrate inhibitors are competitive to the peptide substrate and noncompetitive to the cofactor, suggesting that NatD undergoes an ordered Bi-Bi mechanism. We also demonstrated that these inhibitors are highly specific toward NatD, displaying about 1000-fold selectivity over other closely related acetyltransferases. High-resolution crystal structures of NatD bound to two of these inhibitors revealed the molecular basis for their selectivity and inhibition mechanism, providing a rational path for future inhibitor development.
Organizational Affiliation:
Department of Medicinal Chemistry and Molecular Pharmacology, Purdue Institute for Drug Discovery, Purdue University Center for Cancer Research, Purdue University, West Lafayette, Indiana 47907, United States.