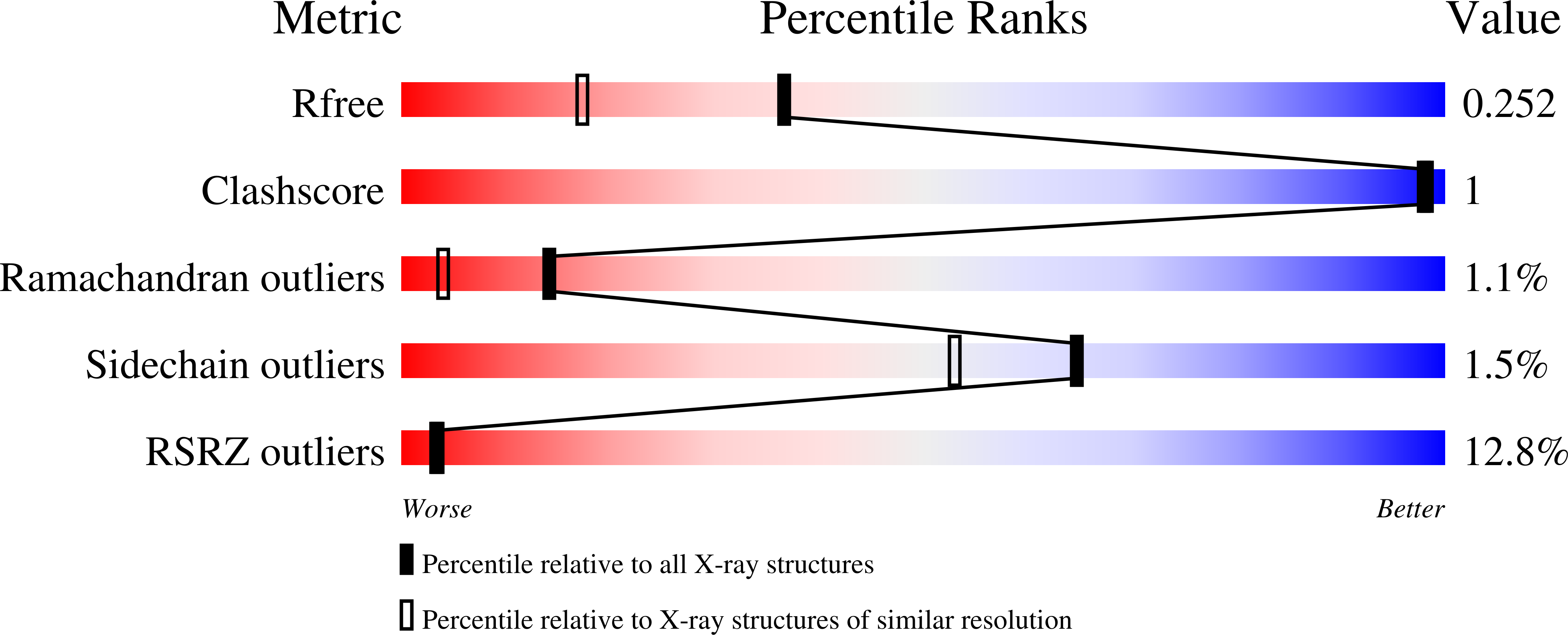
Using yeast surface display to engineer a soluble and crystallizable construct of hematopoietic progenitor kinase 1 (HPK1).
Lau, W.L., Pearce, B., Malakian, H., Rodrigo, I., Xie, D., Gao, M., Marsilio, F., Chang, C., Ruzanov, M., Muckelbauer, J.K., Newitt, J.A., Lipovsek, D., Sheriff, S.(2021) Acta Crystallogr F Struct Biol Commun 77: 22-28
- PubMed: 33439152
- DOI: https://doi.org/10.1107/S2053230X20016015
- Primary Citation of Related Structures:
7KAC - PubMed Abstract:
Hematopoietic progenitor kinase 1 (HPK1) is an intracellular kinase that plays an important role in modulating tumor immune response and thus is an attractive target for drug discovery. Crystallization of the wild-type HPK1 kinase domain has been hampered by poor expression in recombinant systems and poor solubility. In this study, yeast surface display was applied to a library of HPK1 kinase-domain variants in order to select variants with an improved expression level and solubility. The HPK1 variant with the most improved properties contained two mutations, crystallized readily in complex with several small-molecule inhibitors and provided valuable insight to guide structure-based drug design. This work exemplifies the benefit of yeast surface display towards engineering crystallizable proteins and thus enabling structure-based drug discovery.
Organizational Affiliation:
Biologics Discovery, Bristol-Myers Squibb Research and Development, 100 Binney Street, Cambridge, MA 02142, USA.