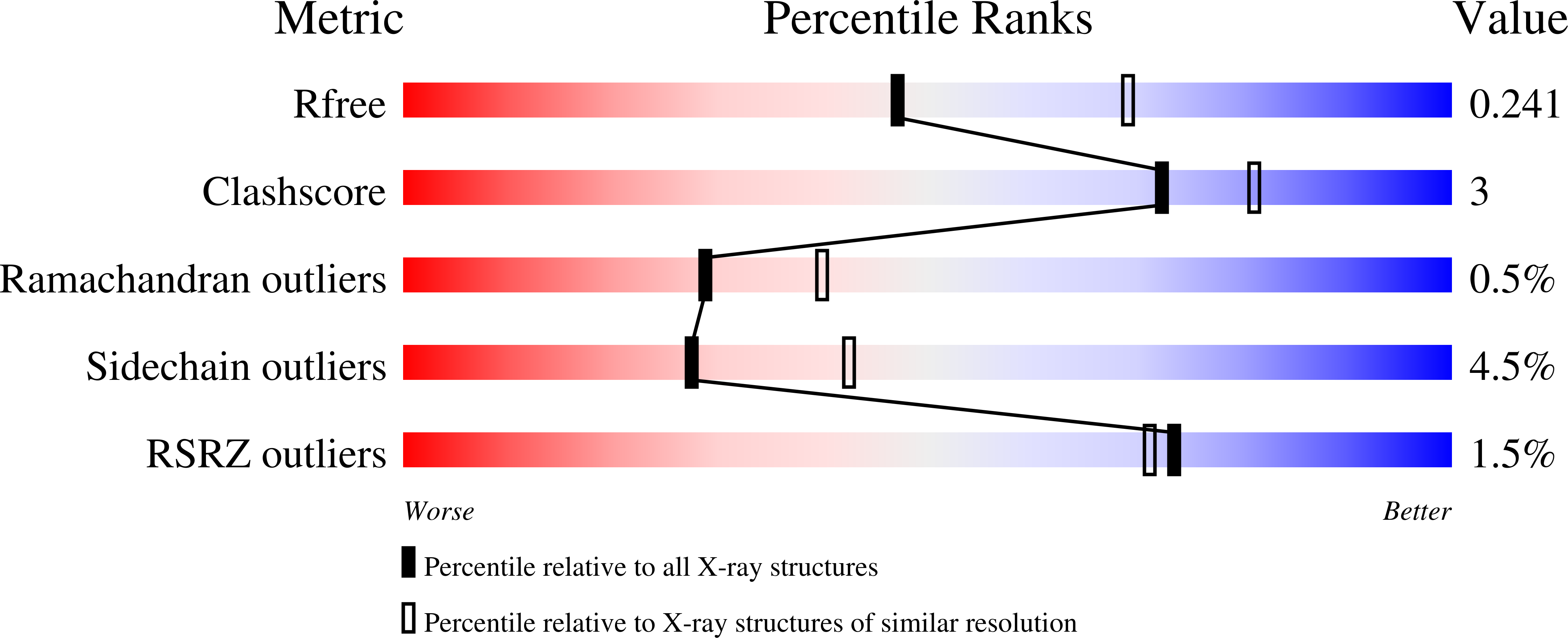
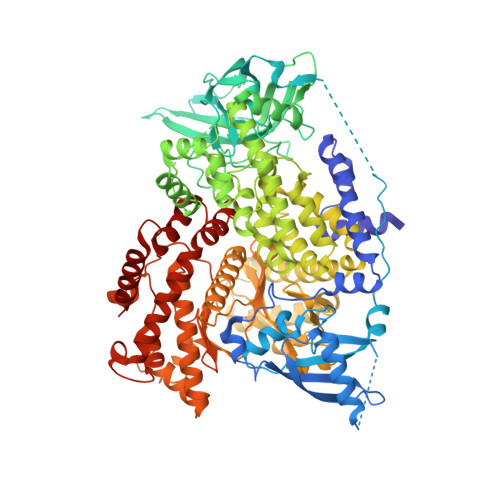
Structure-Based Drug Design and Synthesis of PI3K alpha-Selective Inhibitor (PF-06843195).
Cheng, H., Orr, S.T.M., Bailey, S., Brooun, A., Chen, P., Deal, J.G., Deng, Y.L., Edwards, M.P., Gallego, G.M., Grodsky, N., Huang, B., Jalaie, M., Kaiser, S., Kania, R.S., Kephart, S.E., Lafontaine, J., Ornelas, M.A., Pairish, M., Planken, S., Shen, H., Sutton, S., Zehnder, L., Almaden, C.D., Bagrodia, S., Falk, M.D., Gukasyan, H.J., Ho, C., Kang, X., Kosa, R.E., Liu, L., Spilker, M.E., Timofeevski, S., Visswanathan, R., Wang, Z., Meng, F., Ren, S., Shao, L., Xu, F., Kath, J.C.(2021) J Med Chem 64: 644-661
- PubMed: 33356246
- DOI: https://doi.org/10.1021/acs.jmedchem.0c01652
- Primary Citation of Related Structures:
7K6M, 7K6N, 7K6O, 7K71 - PubMed Abstract:
The phosphoinositide 3-kinase (PI3K)/mammalian target of rapamycin (mTOR) signaling pathway is a frequently dysregulated pathway in human cancer, and PI3Kα is one of the most frequently mutated kinases in human cancer. A PI3Kα-selective inhibitor may provide the opportunity to spare patients the side effects associated with broader inhibition of the class I PI3K family. Here, we describe our efforts to discover a PI3Kα-selective inhibitor by applying structure-based drug design (SBDD) and computational analysis. A novel series of compounds, exemplified by 2,2-difluoroethyl (3 S )-3-{[2'-amino-5-fluoro-2-(morpholin-4-yl)-4,5'-bipyrimidin-6-yl]amino}-3-(hydroxymethyl)pyrrolidine-1-carboxylate ( 1) (PF-06843195), with high PI3Kα potency and unique PI3K isoform and mTOR selectivity were discovered. We describe here the details of the design and synthesis program that lead to the discovery of 1 .
Organizational Affiliation:
La Jolla Laboratories, Pfizer Worldwide Research and Development, 10770 Science Center Drive, San Diego, California 92121, United States.