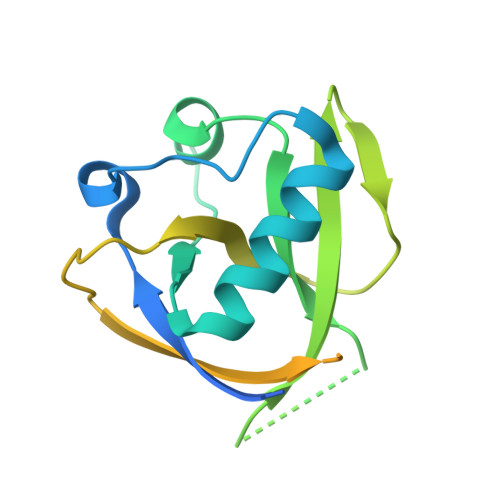
Structural characterization of nonstructural protein 1 from SARS-CoV-2.
Semper, C., Watanabe, N., Savchenko, A.(2021) iScience 24: 101903-101903
- PubMed: 33319167
- DOI: https://doi.org/10.1016/j.isci.2020.101903
- Primary Citation of Related Structures:
7K3N - PubMed Abstract:
Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) is a single-stranded, enveloped RNA virus and the etiological agent of the current coronavirus disease 2019 pandemic. Efficient replication of the virus relies on the activity of nonstructural protein 1 (Nsp1), a major virulence factor shown to facilitate suppression of host gene expression through promotion of host mRNA degradation and interaction with the 40S ribosomal subunit. Here, we report the crystal structure of the globular domain of SARS-CoV-2 Nsp1, encompassing residues 13 to 127, at a resolution of 1.65 Å. Our structure features a six-stranded, capped β-barrel motif similar to Nsp1 from SARS-CoV and reveals how variations in amino acid sequence manifest as distinct structural features. Combining our high-resolution crystal structure with existing data on the C-terminus of Nsp1 from SARS-CoV-2, we propose a model of the full-length protein. Our results provide insight into the molecular structure of a major pathogenic determinant of SARS-CoV-2.
- Department of Microbiology, Immunology and Infectious Disease, University of Calgary, HSC B724 3330 Hospital Drive NW, Calgary, Alberta, T2N 4N1, Canada.
Organizational Affiliation: