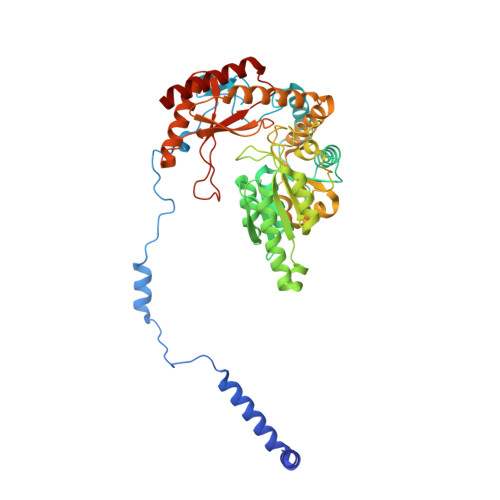
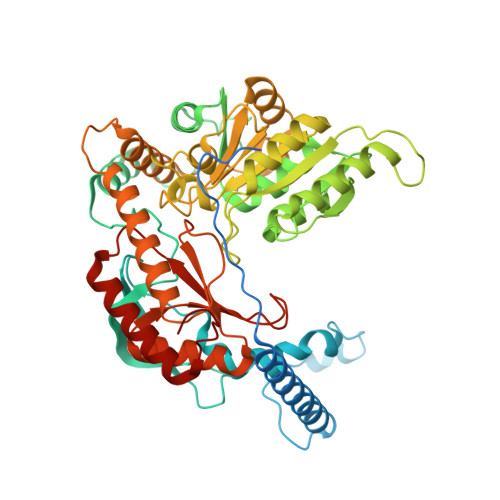
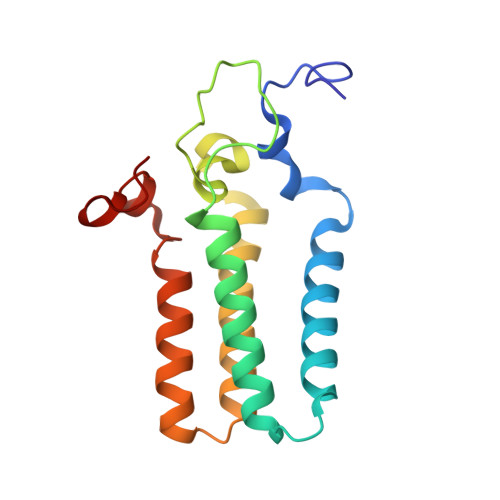
Structural insights into the regulation of human serine palmitoyltransferase complexes.
Wang, Y., Niu, Y., Zhang, Z., Gable, K., Gupta, S.D., Somashekarappa, N., Han, G., Zhao, H., Myasnikov, A.G., Kalathur, R.C., Dunn, T.M., Lee, C.H.(2021) Nat Struct Mol Biol 28: 240-248
- PubMed: 33558761
- DOI: https://doi.org/10.1038/s41594-020-00551-9
- Primary Citation of Related Structures:
7K0I, 7K0J, 7K0K, 7K0L, 7K0M, 7K0N, 7K0O, 7K0P, 7K0Q - PubMed Abstract:
Sphingolipids are essential lipids in eukaryotic membranes. In humans, the first and rate-limiting step of sphingolipid synthesis is catalyzed by the serine palmitoyltransferase holocomplex, which consists of catalytic components (SPTLC1 and SPTLC2) and regulatory components (ssSPTa and ORMDL3). However, the assembly, substrate processing and regulation of the complex are unclear. Here, we present 8 cryo-electron microscopy structures of the human serine palmitoyltransferase holocomplex in various functional states at resolutions of 2.6-3.4 Å. The structures reveal not only how catalytic components recognize the substrate, but also how regulatory components modulate the substrate-binding tunnel to control enzyme activity: ssSPTa engages SPTLC2 and shapes the tunnel to determine substrate specificity. ORMDL3 blocks the tunnel and competes with substrate binding through its amino terminus. These findings provide mechanistic insights into sphingolipid biogenesis governed by the serine palmitoyltransferase complex.
Organizational Affiliation:
Department of Structural Biology, St. Jude Children's Research Hospital, Memphis, TN, USA.