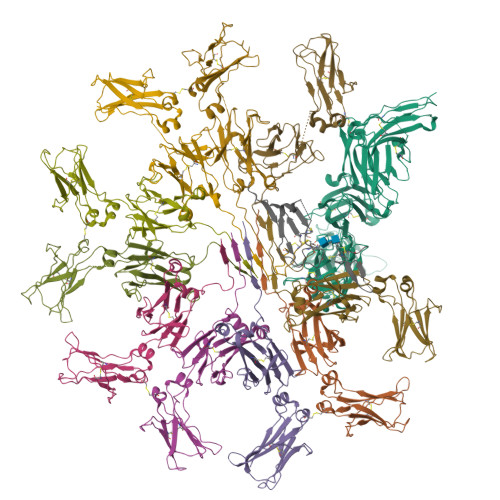
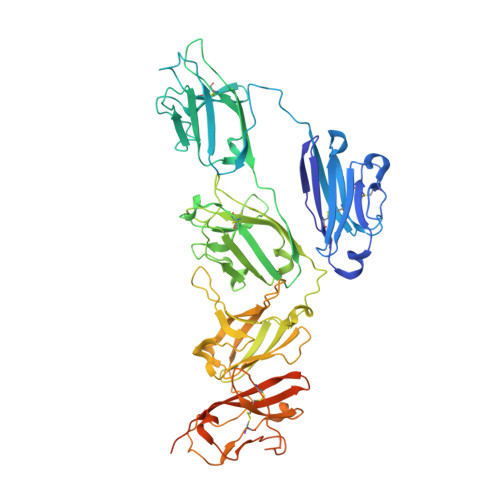
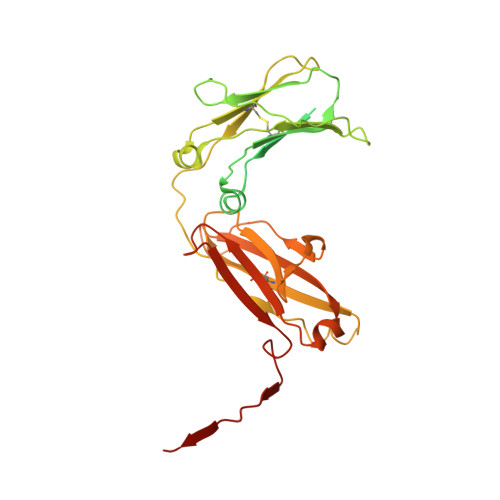
Structure of the human secretory immunoglobulin M core.
Kumar, N., Arthur, C.P., Ciferri, C., Matsumoto, M.L.(2021) Structure 29: 564-571.e3
- PubMed: 33513362
- DOI: https://doi.org/10.1016/j.str.2021.01.002
- Primary Citation of Related Structures:
7K0C - PubMed Abstract:
Immunoglobulins (Ig) A and M are the only human antibodies that form oligomers and undergo transcytosis to mucosal secretions via the polymeric Ig receptor (pIgR). When complexed with the J-chain (JC) and the secretory component (SC) of pIgR, secretory IgA and IgM (sIgA and sIgM) play critical roles in host-pathogen defense. Recently, we determined the structure of sIgA-Fc which elucidated the mechanism of polymeric IgA assembly and revealed an extensive binding interface between IgA-Fc, JC, and SC. Despite low sequence identity shared with IgA-Fc, IgM-Fc also undergoes JC-mediated assembly and binds pIgR. Here, we report the structure of sIgM-Fc and carryout a systematic comparison to sIgA-Fc. Our structural analysis reveals a remarkably conserved mechanism of JC-templated oligomerization and SC recognition of both IgM and IgA through a highly conserved network of interactions. These studies reveal the structurally conserved features of sIgM and sIgA required for function in mucosal immunity.
Organizational Affiliation:
Department of Structural Biology, Genentech, 1 DNA Way, South San Francisco, CA 94080, USA.