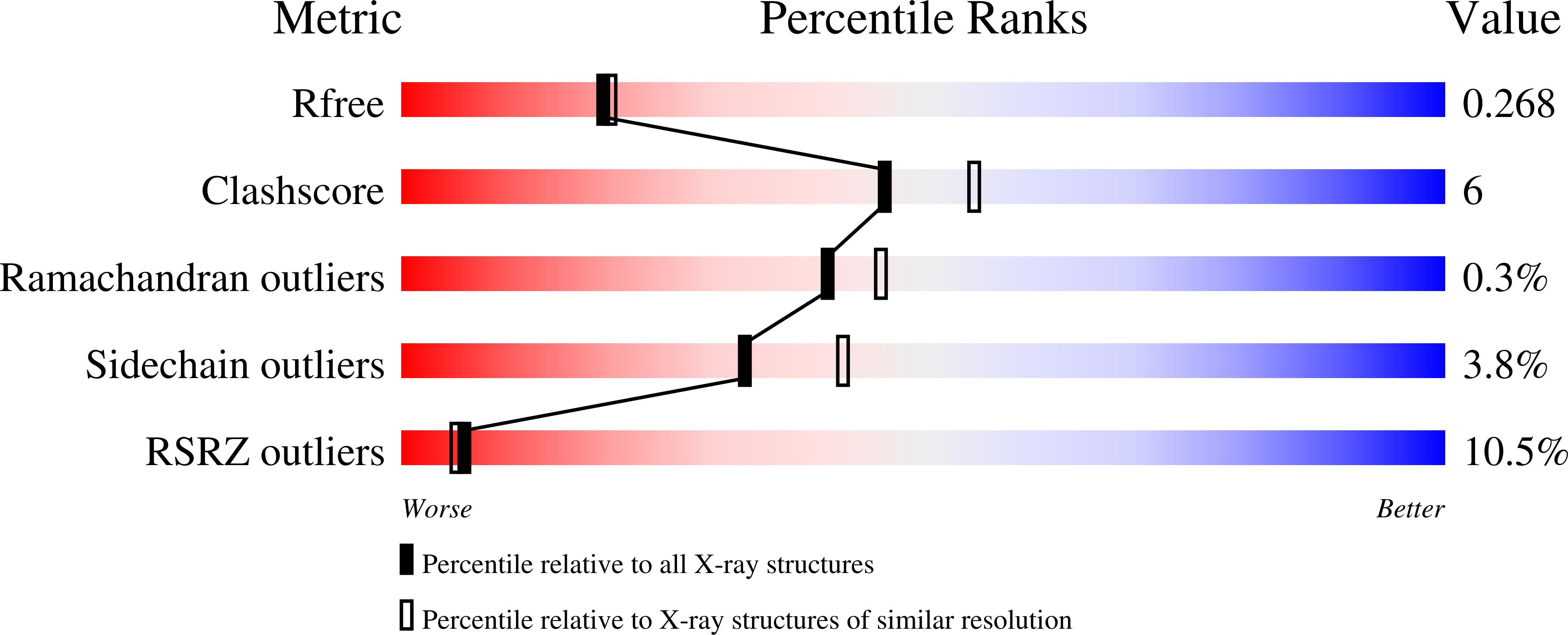
Molecular basis for cooperative binding and synergy of ATP-site and allosteric EGFR inhibitors
Beyett, T.S., To, C., Heppner, D.E., Rana, J.K., Schmoker, A.M., Jang, J., De Clercq, D.J.H., Gomez, G., Scott, D.A., Gray, N.S., Janne, P.A., Eck, M.J.(2022) Nat Commun 13: 2530