Development of 2,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one inhibitors of aldehyde dehydrogenase 1A (ALDH1A) as potential adjuncts to ovarian cancer chemotherapy.
Huddle, B.C., Grimley, E., Chtcherbinine, M., Buchman, C.D., Takahashi, C., Debnath, B., McGonigal, S.C., Mao, S., Li, S., Felton, J., Pan, S., Wen, B., Sun, D., Neamati, N., Buckanovich, R.J., Hurley, T.D., Larsen, S.D.(2020) Eur J Med Chem 211: 113060-113060
- PubMed: 33341649
- DOI: https://doi.org/10.1016/j.ejmech.2020.113060
- Primary Citation of Related Structures:
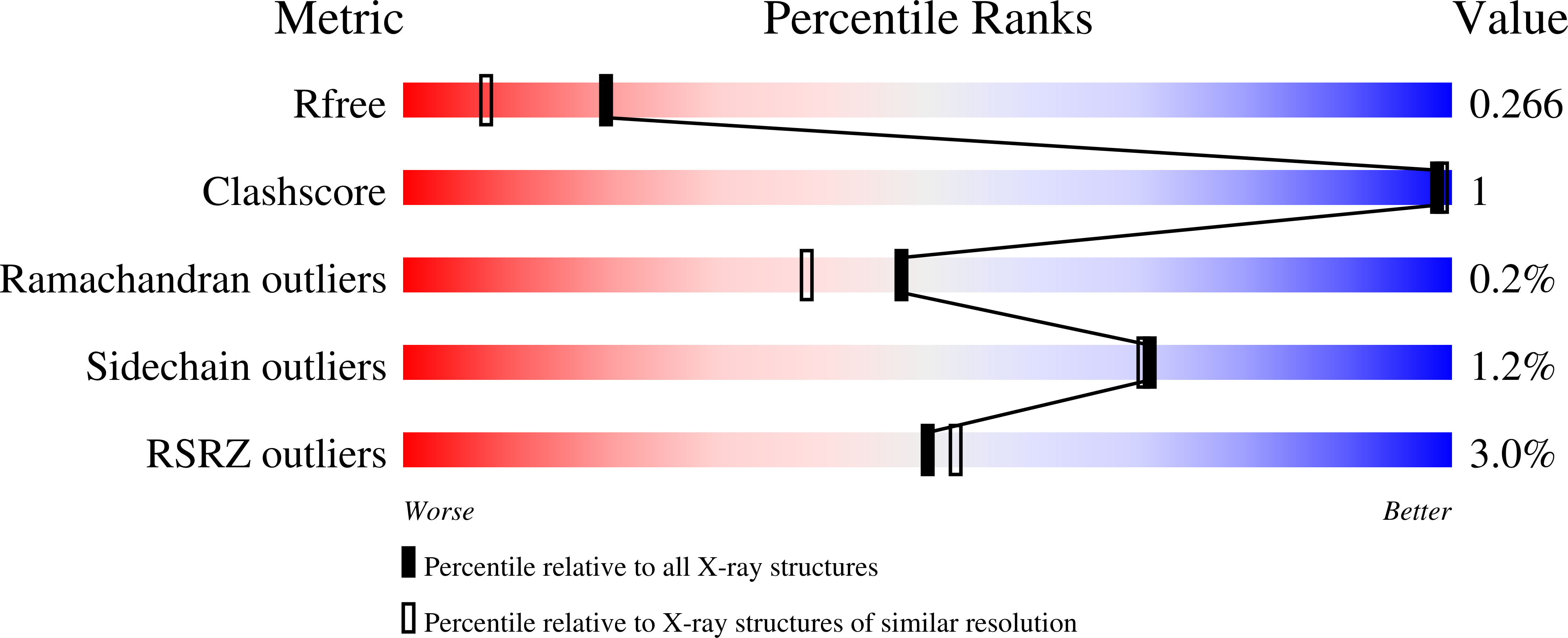
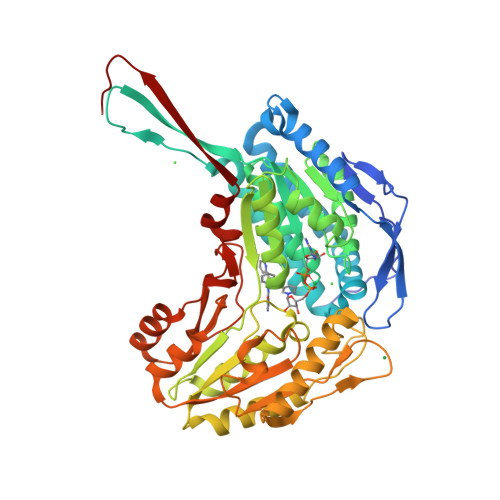
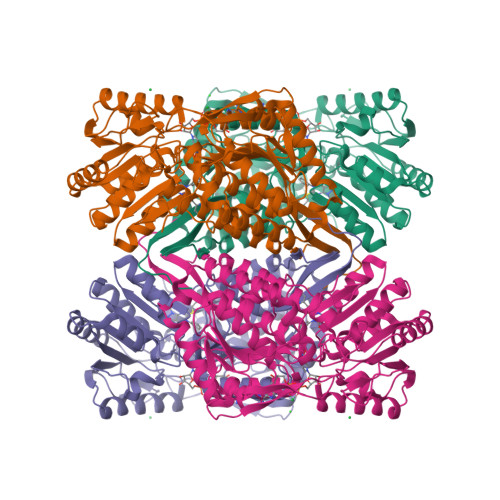
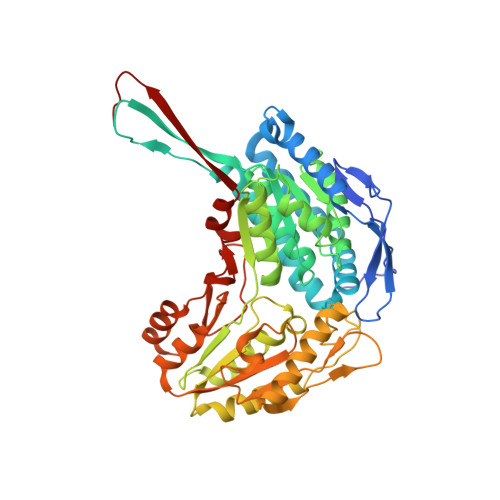
7JWS, 7JWT, 7JWU, 7JWV, 7JWW - PubMed Abstract:
There is strong evidence that inhibition of one or more Aldehyde Dehydrogenase 1A (ALDH1A) isoforms may be beneficial in chemotherapy-resistant ovarian cancer and other tumor types. While many previous efforts have focused on development of ALDH1A1 selective inhibitors, the most deadly ovarian cancer subtype, high-grade serous (HGSOC), exhibits elevated expression of ALDH1A3. Herein, we report continued development of pan-ALDH1A inhibitors to assess whether broad spectrum ALDH1A inhibition is an effective adjunct to chemotherapy in this critical tumor subtype. Optimization of the CM39 scaffold, aided by metabolite ID and several new ALDH1A1 crystal structures, led to improved biochemical potencies, improved cellular ALDH inhibition in HGSOC cell lines, and substantial improvements in microsomal stability culminating in orally bioavailable compounds. We demonstrate that two compounds 68 and 69 are able to synergize with chemotherapy in a resistant cell line and patient-derived HGSOC tumor spheroids, indicating their suitability for future in vivo proof of concept experiments.
Organizational Affiliation:
Department of Medicinal Chemistry, College of Pharmacy, University of Michigan, Ann Arbor, MI, 48109, USA.