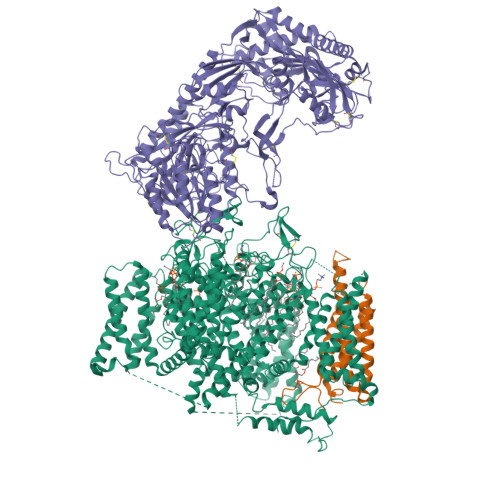
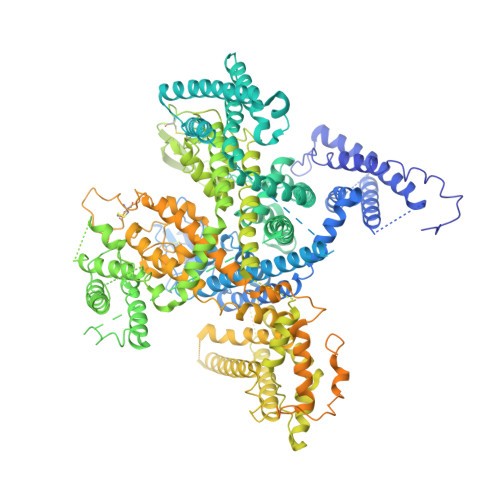
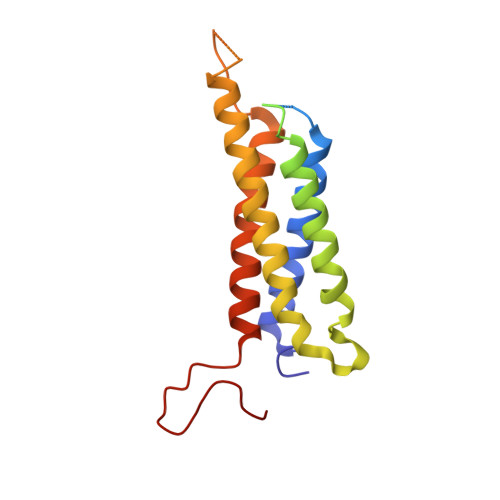
Structural Basis of the Modulation of the Voltage-Gated Calcium Ion Channel Ca v 1.1 by Dihydropyridine Compounds*.
Gao, S., Yan, N.(2021) Angew Chem Int Ed Engl 60: 3131-3137
- PubMed: 33125829
- DOI: https://doi.org/10.1002/anie.202011793
- Primary Citation of Related Structures:
7JPK, 7JPL, 7JPV, 7JPW, 7JPX - PubMed Abstract:
1,4-Dihydropyridines (DHP), the most commonly used antihypertensives, function by inhibiting the L-type voltage-gated Ca 2+ (Ca v ) channels. DHP compounds exhibit chirality-specific antagonistic or agonistic effects. The structure of rabbit Ca v 1.1 bound to an achiral drug nifedipine reveals the general binding mode for DHP drugs, but the molecular basis for chiral specificity remained elusive. Herein, we report five cryo-EM structures of nanodisc-embedded Ca v 1.1 in the presence of the bestselling drug amlodipine, a DHP antagonist (R)-(+)-Bay K8644, and a titration of its agonistic enantiomer (S)-(-)-Bay K8644 at resolutions of 2.9-3.4 Å. The amlodipine-bound structure reveals the molecular basis for the high efficacy of the drug. All structures with the addition of the Bay K8644 enantiomers exhibit similar inactivated conformations, suggesting that (S)-(-)-Bay K8644, when acting as an agonist, is insufficient to lock the activated state of the channel for a prolonged duration.
Organizational Affiliation:
Department of Molecular Biology, Princeton University, Princeton, NJ, 08544, USA.