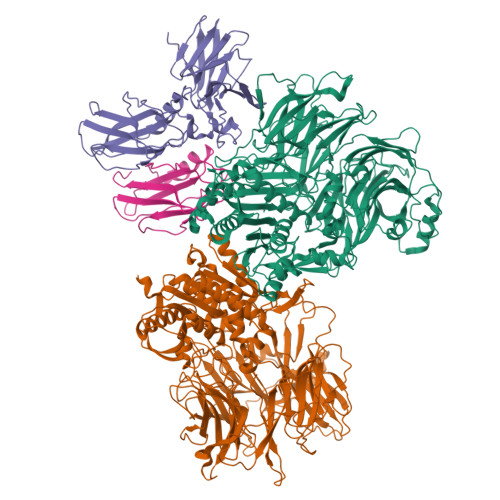
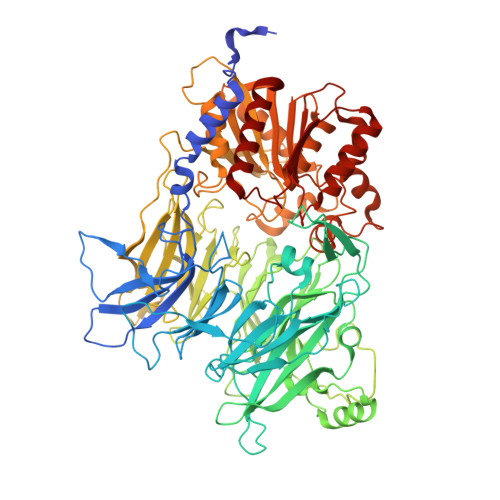
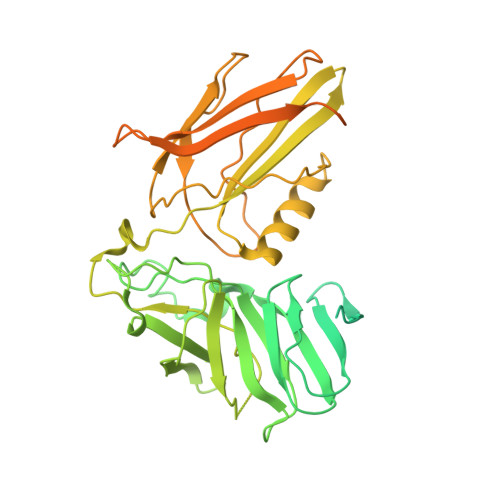
Dipeptidyl peptidase 9 sets a threshold for CARD8 inflammasome formation by sequestering its active C-terminal fragment.
Sharif, H., Hollingsworth, L.R., Griswold, A.R., Hsiao, J.C., Wang, Q., Bachovchin, D.A., Wu, H.(2021) Immunity 54: 1392
- PubMed: 34019797
- DOI: https://doi.org/10.1016/j.immuni.2021.04.024
- Primary Citation of Related Structures:
7JKQ, 7JN7 - PubMed Abstract:
CARD8 detects intracellular danger signals and forms a caspase-1 activating inflammasome. Like the related inflammasome sensor NLRP1, CARD8 autoprocesses into noncovalently associated N-terminal (NT) and C-terminal (CT) fragments and binds the cellular dipeptidyl peptidases DPP8 and 9 (DPP8/9). Certain danger-associated signals, including the DPP8/9 inhibitor Val-boroPro (VbP) and HIV protease, induce proteasome-mediated NT degradation and thereby liberate the inflammasome-forming CT. Here, we report cryoelectron microscopy (cryo-EM) structures of CARD8 bound to DPP9, revealing a repressive ternary complex consisting of DPP9, full-length CARD8, and CARD8-CT. Unlike NLRP1-CT, CARD8-CT does not interact with the DPP8/9 active site and is not directly displaced by VbP. However, larger DPP8/9 active-site probes can directly weaken this complex in vitro, and VbP itself nevertheless appears to disrupt this complex, perhaps indirectly, in cells. Thus, DPP8/9 inhibitors can activate the CARD8 inflammasome by promoting CARD8 NT degradation and by weakening ternary complex stability.
Organizational Affiliation:
Department of Biological Chemistry and Molecular Pharmacology, Harvard Medical School, Boston, MA 02115, USA; Program in Cellular and Molecular Medicine, Boston Children's Hospital, Boston, MA 02115, USA.