The 0.84 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with palmitoleic acid
Sugiyama, S., Kakinouchi, K., Matsuoka, S., Tsuchikawa, H., Sonoyama, M., Inoue, Y., Hayashi, F., Murata, M.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Fatty acid-binding protein, heart | 133 | Homo sapiens | Mutation(s): 0 Gene Names: FABP3, FABP11, MDGI | 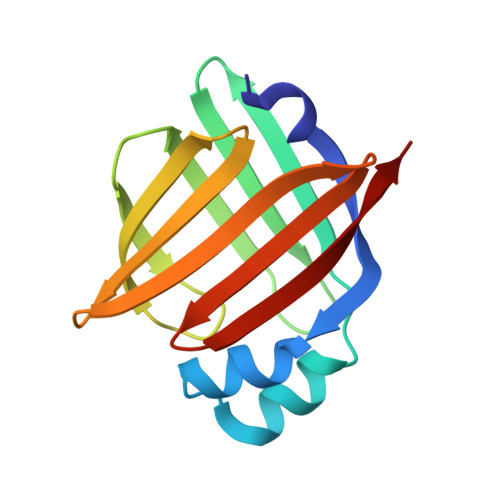 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P05413 (Homo sapiens) Explore P05413 Go to UniProtKB: P05413 | |||||
PHAROS: P05413 GTEx: ENSG00000121769 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P05413 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| P6G (Subject of Investigation/LOI) Query on P6G | C [auth A], D [auth A] | HEXAETHYLENE GLYCOL C12 H26 O7 IIRDTKBZINWQAW-UHFFFAOYSA-N |  | ||
| PAM (Subject of Investigation/LOI) Query on PAM | B [auth A] | PALMITOLEIC ACID C16 H30 O2 SECPZKHBENQXJG-FPLPWBNLSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 54.536 | α = 90 |
| b = 69.298 | β = 90 |
| c = 33.818 | γ = 90 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| HKL-2000 | data reduction |
| HKL-2000 | data scaling |
| MOLREP | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Japan Science and Technology | Japan | JPMJTM19DC |
| Japan Science and Technology | Japan | JPMJER1005 |
| Japan Society for the Promotion of Science (JSPS) | Japan | 19K06588 |
| Other private | Jamaica | 09-003-005 |