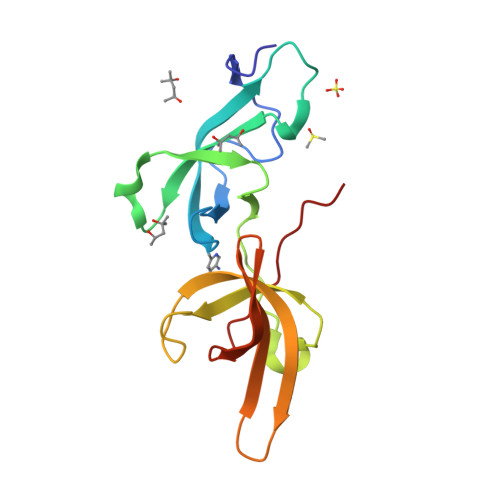
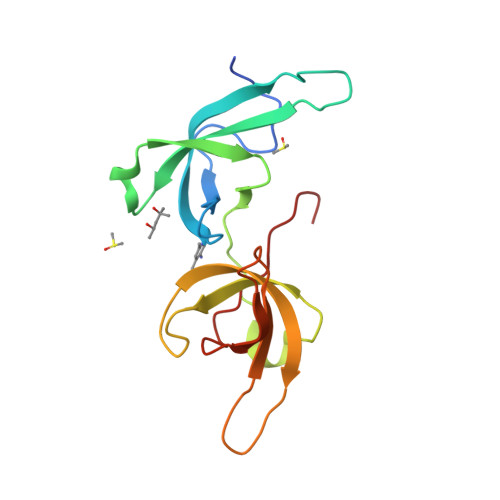
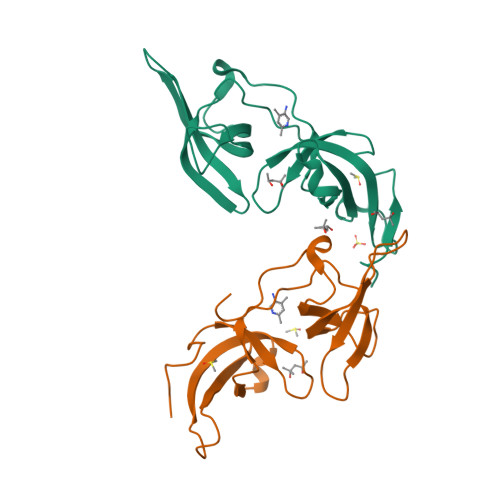Structure-based screening combined with computational and biochemical analyses identified the inhibitor targeting the binding of DNA Ligase 1 to UHRF1.
Kori, S., Shibahashi, Y., Ekimoto, T., Nishiyama, A., Yoshimi, S., Yamaguchi, K., Nagatoishi, S., Ohta, M., Tsumoto, K., Nakanishi, M., Defossez, P.A., Ikeguchi, M., Arita, K.(2021) Bioorg Med Chem 52: 116500-116500
- PubMed: 34801826
- DOI: https://doi.org/10.1016/j.bmc.2021.116500
- Primary Citation of Related Structures:
7FB7 - PubMed Abstract:
The accumulation of epigenetic alterations is one of the major causes of tumorigenesis. Aberrant DNA methylation patterns cause genome instability and silencing of tumor suppressor genes in various types of tumors. Therefore, drugs that target DNA methylation-regulating factors have great potential for cancer therapy. Ubiquitin-like containing PHD and RING finger domain 1 (UHRF1) is an essential factor for DNA methylation maintenance. UHRF1 is overexpressed in various cancer cells and down-regulation of UHRF1 in these cells reactivates the expression of tumor suppressor genes, thus UHRF1 is a promising target for cancer therapy. We have previously shown that interaction between the tandem Tudor domain (TTD) of UHRF1 and DNA ligase 1 (LIG1) di/trimethylated on Lys126 plays a key role in the recruitment of UHRF1 to replication sites and replication-coupled DNA methylation maintenance. An arginine binding cavity (Arg-binding cavity) of the TTD is essential for LIG1 interaction, thus the development of inhibitors that target the Arg-binding cavity could potentially repress UHRF1 function in cancer cells. To develop such an inhibitor, we performed in silico screening using not only static but also dynamic metrics based on all-atom molecular dynamics simulations, resulting in efficient identification of 5-amino-2,4-dimethylpyridine (5A-DMP) as a novel TTD-binding compound. Crystal structure of the TTD in complex with 5A-DMP revealed that the compound stably bound to the Arg-binding cavity of the TTD. Furthermore, 5A-DMP inhibits the full-length UHRF1:LIG1 interaction in Xenopus egg extracts. Our study uncovers a UHRF1 inhibitor which can be the basis of future experiments for cancer therapy.
Organizational Affiliation:
Structural Biology Laboratory, Graduate School of Medical Life Science, Yokohama City University, 1-7-29, Suehiro-cho, Tsurumi-ku, Yokohama, Kanagawa 230-0045, Japan.