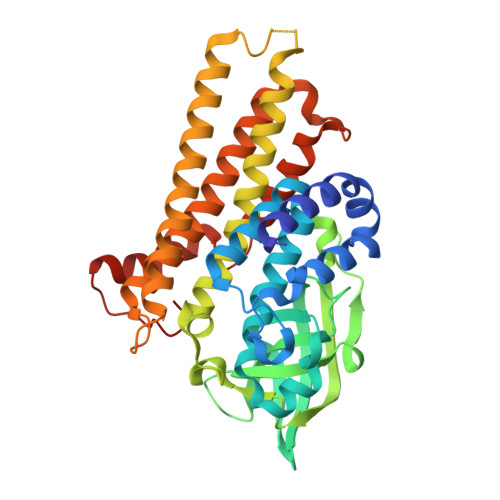
Omics analysis of Mycobacterium tuberculosis isolates uncovers Rv3094c, an ethionamide metabolism-associated gene.
Wan, L., Hu, P., Zhang, L., Wang, Z.X., Fleming, J., Ni, B., Luo, J., Guan, C.X., Bai, L., Tan, Y., Liu, H., Li, N., Xiao, T., Bai, H., Zhang, Y.A., Zhang, X.E., Wan, K., Bi, L., Ouyang, S., Zhang, H.(2023) Commun Biol 6: 156-156
- PubMed: 36750726
- DOI: https://doi.org/10.1038/s42003-023-04433-w
- Primary Citation of Related Structures:
7F70, 7F72, 7F74 - PubMed Abstract:
Global control of the tuberculosis epidemic is threatened by increasing prevalence of drug resistant M. tuberculosis isolates. Many genome-wide studies focus on SNP-associated drug resistance mechanisms, but drug resistance in 5-30% of M. tuberculosis isolates (varying with antibiotic) appears unrelated to reported SNPs, and alternative drug resistance mechanisms involving variation in gene/protein expression are not well-studied. Here, using an omics approach, we identify 388 genes with lineage-related differential expression and 68 candidate drug resistance-associated gene pairs/clusters in 11 M. tuberculosis isolates (variable lineage/drug resistance profiles). Structural, mutagenesis, biochemical and bioinformatic studies on Rv3094c from the Rv3093c-Rv3095 gene cluster, a gene cluster selected for further investigation as it contains a putative monooxygenase/repressor pair and is associated with ethionamide resistance, provide insights on its involvement in ethionamide sulfoxidation, the initial step in its activation. Analysis of the structure of Rv3094c and its complex with ethionamide and flavin mononucleotide, to the best of our knowledge the first structures of an enzyme involved in ethionamide activation, identify key residues in the flavin mononucleotide and ethionamide binding pockets of Rv3094c, and F221, a gate between flavin mononucleotide and ethionamide allowing their interaction to complete the sulfoxidation reaction. Our work broadens understanding of both lineage- and drug resistance-associated gene/protein expression perturbations and identifies another player in mycobacterial ethionamide metabolism.
Organizational Affiliation:
Key Laboratory of RNA Biology, Institute of Biophysics, Chinese Academy of Sciences, Beijing, 100101, China.