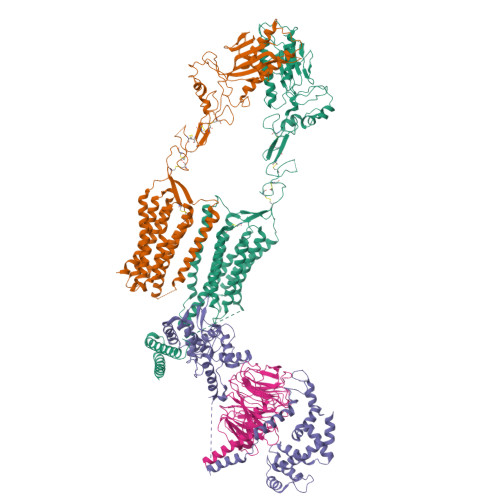
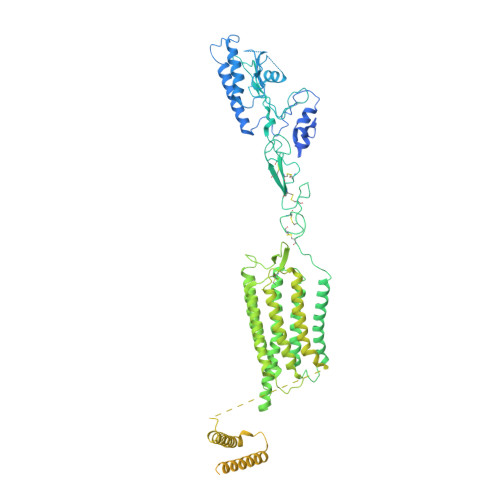
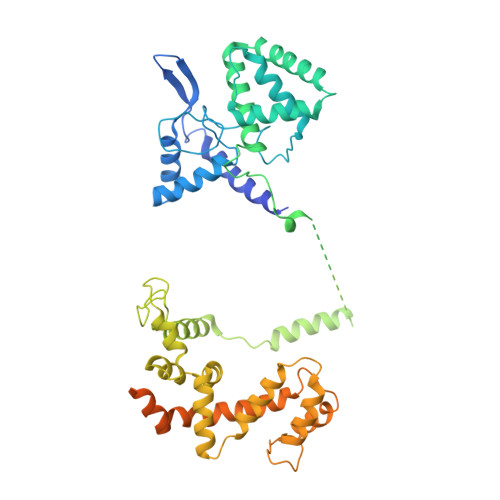
Structure of the class C orphan GPCR GPR158 in complex with RGS7-G beta 5.
Jeong, E., Kim, Y., Jeong, J., Cho, Y.(2021) Nat Commun 12: 6805-6805
- PubMed: 34815401
- DOI: https://doi.org/10.1038/s41467-021-27147-1
- Primary Citation of Related Structures:
7EWL, 7EWP, 7EWR - PubMed Abstract:
GPR158, a class C orphan GPCR, functions in cognition, stress-induced mood control, and synaptic development. Among class C GPCRs, GPR158 is unique as it lacks a Venus flytrap-fold ligand-binding domain and terminates Gαi/o protein signaling through the RGS7-Gβ5 heterodimer. Here, we report the cryo-EM structures of GPR158 alone and in complex with one or two RGS7-Gβ5 heterodimers. GPR158 dimerizes through Per-Arnt-Sim-fold extracellular and transmembrane (TM) domains connected by an epidermal growth factor-like linker. The TM domain (TMD) reflects both inactive and active states of other class C GPCRs: a compact intracellular TMD, conformations of the two intracellular loops (ICLs) and the TMD interface formed by TM4/5. The ICL2, ICL3, TM3, and first helix of the cytoplasmic coiled-coil provide a platform for the DHEX domain of one RGS7 and the second helix recruits another RGS7. The unique features of the RGS7-binding site underlie the selectivity of GPR158 for RGS7.
Organizational Affiliation:
Department of Life Science, Pohang University of Science and Technology, Pohang, Republic of Korea.