Functional Characterization and Structural Insights Into Stereoselectivity of Pulegone Reductase in Menthol Biosynthesis.
Liu, C., Gao, Q., Shang, Z., Liu, J., Zhou, S., Dang, J., Liu, L., Lange, I., Srividya, N., Lange, B.M., Wu, Q., Lin, W.(2021) Front Plant Sci 12: 780970-780970
- PubMed: 34917113
- DOI: https://doi.org/10.3389/fpls.2021.780970
- Primary Citation of Related Structures:
7EQL - PubMed Abstract:
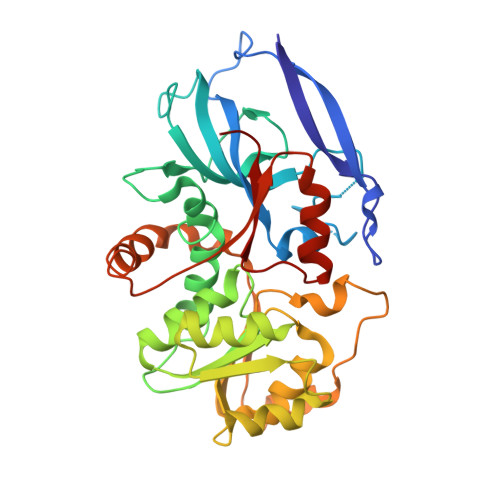
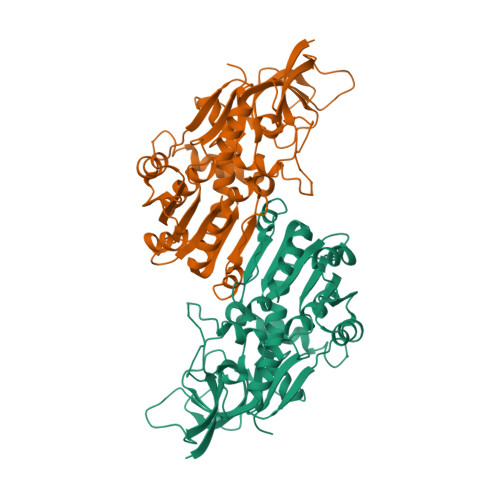
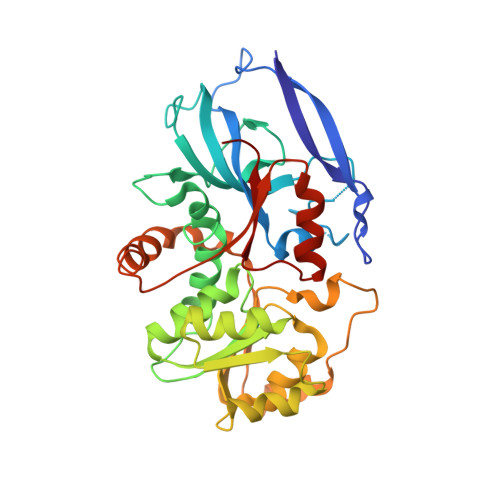
Monoterpenoids are the main components of plant essential oils and the active components of some traditional Chinese medicinal herbs like Mentha haplocalyx Briq ., Nepeta tenuifolia Briq ., Perilla frutescens (L.) Britt and Pogostemin cablin (Blanco) Benth. Pulegone reductase is the key enzyme in the biosynthesis of menthol and is required for the stereoselective reduction of the Δ 2,8 double bond of pulegone to produce the major intermediate menthone, thus determining the stereochemistry of menthol. However, the structural basis and mechanism underlying the stereoselectivity of pulegone reductase remain poorly understood. In this study, we characterized a novel (-)-pulegone reductase from Nepeta tenuifolia ( Nt PR), which can catalyze (-)-pulegone to (+)-menthone and (-)-isomenthone through our RNA-seq, bioinformatic analysis in combination with in vitro enzyme activity assay, and determined the structure of (+)-pulegone reductase from M. piperita ( Mp PR) by using X-ray crystallography, molecular modeling and docking, site-directed mutagenesis, molecular dynamics simulations, and biochemical analysis. We identified and validated the critical residues in the crystal structure of Mp PR involved in the binding of the substrate pulegone. We also further identified that residues Leu56, Val282, and Val284 determine the stereoselectivity of the substrate pulegone, and mainly contributes to the product stereoselectivity. This work not only provides a starting point for the understanding of stereoselectivity of pulegone reductases, but also offers a basis for the engineering of menthone/menthol biosynthetic enzymes to achieve high-titer, industrial-scale production of enantiomerically pure products.
Organizational Affiliation:
School of Pharmacy, Nanjing University of Chinese Medicine, Nanjing, China.