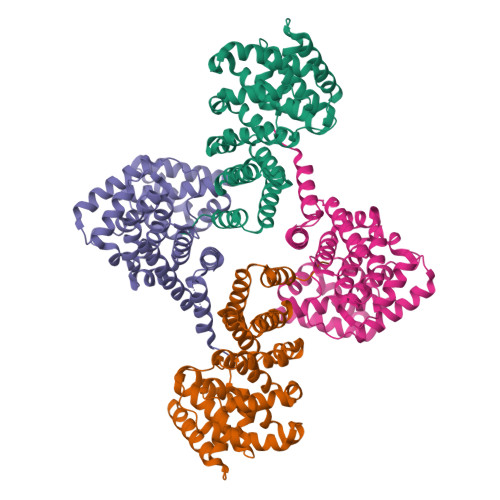
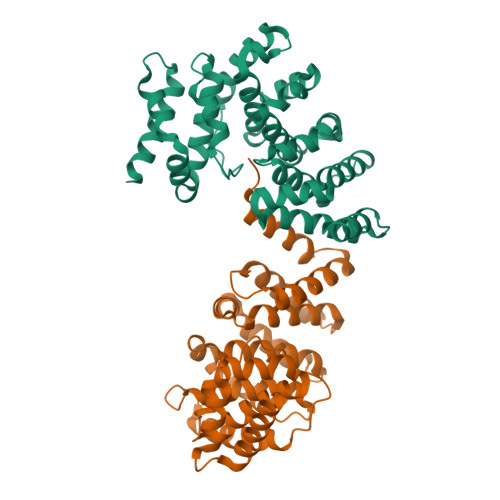
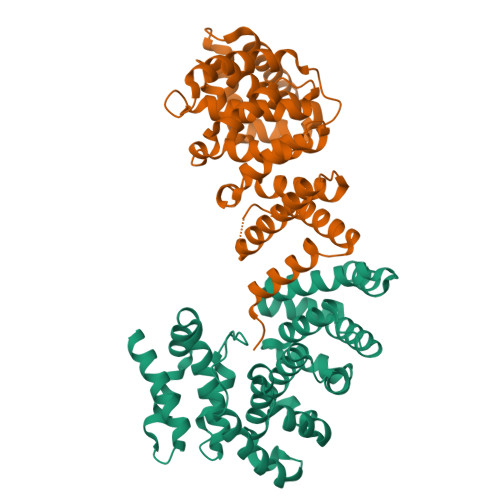
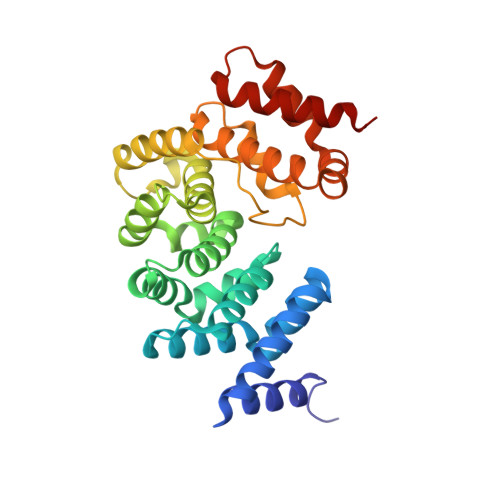
Molecular basis for recognition of Gly/N-degrons by CRL2 ZYG11B and CRL2 ZER1 .
Yan, X., Li, Y., Wang, G., Zhou, Z., Song, G., Feng, Q., Zhao, Y., Mi, W., Ma, Z., Dong, C.(2021) Mol Cell 81: 3262-3274.e3
- PubMed: 34214466
- DOI: https://doi.org/10.1016/j.molcel.2021.06.010
- Primary Citation of Related Structures:
7EP0, 7EP1, 7EP2, 7EP3, 7EP4, 7EP5 - PubMed Abstract:
N-degron pathways are a set of proteolytic systems that target the N-terminal destabilizing residues of substrates for proteasomal degradation. Recently, the Gly/N-degron pathway has been identified as a new branch of the N-degron pathway. The N-terminal glycine degron (Gly/N-degron) is recognized by ZYG11B and ZER1, the substrate receptors of the Cullin 2-RING E3 ubiquitin ligase (CRL2). Here we present the crystal structures of ZYG11B and ZER1 bound to various Gly/N-degrons. The structures reveal that ZYG11B and ZER1 utilize their armadillo (ARM) repeats forming a deep and narrow cavity to engage mainly the first four residues of Gly/N-degrons. The α-amino group of the Gly/N-degron is accommodated in an acidic pocket by five conserved hydrogen bonds. These structures, together with biochemical studies, decipher the molecular basis for the specific recognition of the Gly/N-degron by ZYG11B and ZER1, providing key information for future structure-based chemical probe design.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, The Province and Ministry Co-sponsored Collaborative Innovation Center for Medical Epigenetics, Key Laboratory of Immune Microenvironment and Disease (Ministry of Education), School of Basic Medical Sciences, Tianjin Medical University, Tianjin 300070, China.