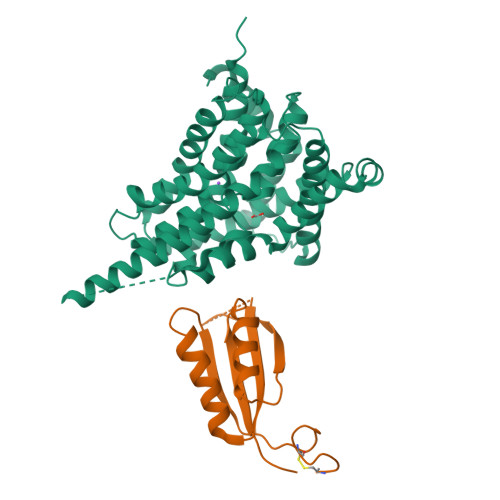
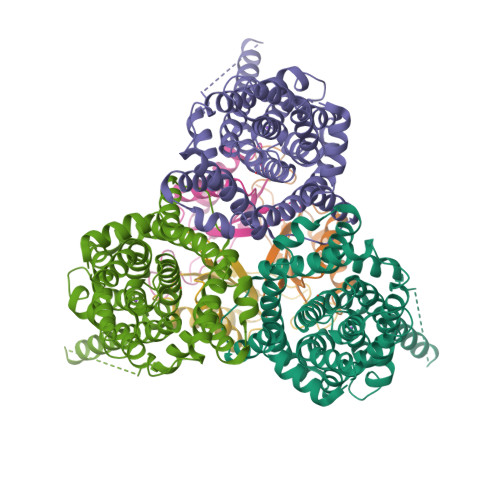
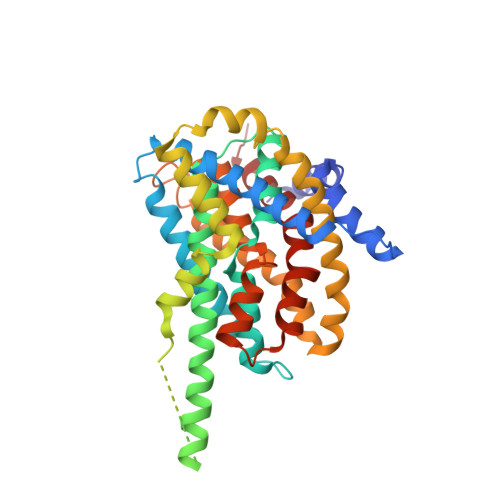
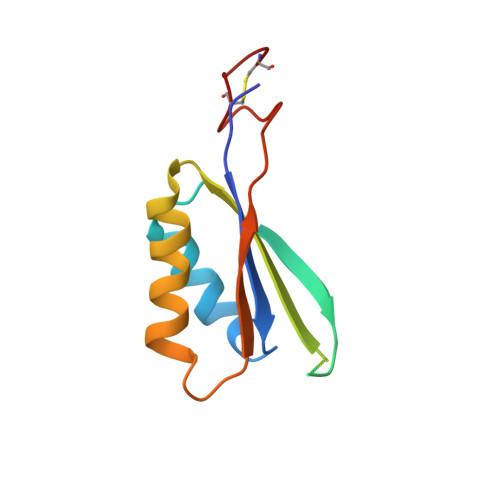
Molecular mechanism underlying transport and allosteric inhibition of bicarbonate transporter SbtA.
Fang, S., Huang, X., Zhang, X., Zhang, M., Hao, Y., Guo, H., Liu, L.N., Yu, F., Zhang, P.(2021) Proc Natl Acad Sci U S A 118
- PubMed: 34031249
- DOI: https://doi.org/10.1073/pnas.2101632118
- Primary Citation of Related Structures:
7EGK, 7EGL - PubMed Abstract:
SbtA is a high-affinity, sodium-dependent bicarbonate transporter found in the cyanobacterial CO 2 -concentrating mechanism (CCM). SbtA forms a complex with SbtB, while SbtB allosterically regulates the transport activity of SbtA by binding with adenyl nucleotides. The underlying mechanism of transport and regulation of SbtA is largely unknown. In this study, we report the three-dimensional structures of the cyanobacterial Synechocystis sp. PCC 6803 SbtA-SbtB complex in both the presence and absence of HCO 3 - and/or AMP at 2.7 Å and 3.2 Å resolution. An analysis of the inward-facing state of the SbtA structure reveals the HCO 3 - /Na + binding site, providing evidence for the functional unit as a trimer. A structural comparison found that SbtA adopts an elevator mechanism for bicarbonate transport. A structure-based analysis revealed that the allosteric inhibition of SbtA by SbtB occurs mainly through the T-loop of SbtB, which binds to both the core domain and the scaffold domain of SbtA and locks it in an inward-facing state. T-loop conformation is stabilized by the AMP molecules binding at the SbtB trimer interfaces and may be adjusted by other adenyl nucleotides. The unique regulatory mechanism of SbtA by SbtB makes it important to study inorganic carbon uptake systems in CCM, which can be used to modify photosynthesis in crops.
Organizational Affiliation:
National Key Laboratory of Plant Molecular Genetics, Center for Excellence in Molecular Plant Sciences, Institute of Plant Physiology and Ecology, Chinese Academy of Sciences, Shanghai 200032, China.