Chiral deaza-coelenterazine analogs for probing a substrate-binding site in the Ca2+-binding photoprotein aequorin.
Inouye, S., Sumida, Y., Tomabechi, Y., Taguchi, J., Shirouzu, M., Hosoya, T.(2021) PLoS One 16: e0251743-e0251743
- PubMed: 34115795
- DOI: https://doi.org/10.1371/journal.pone.0251743
- Primary Citation of Related Structures:
7EG2, 7EG3 - PubMed Abstract:
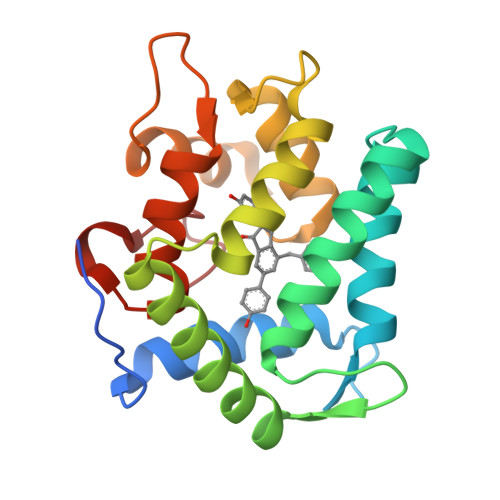
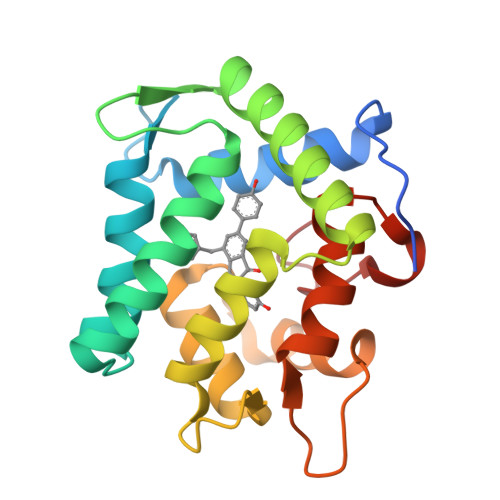
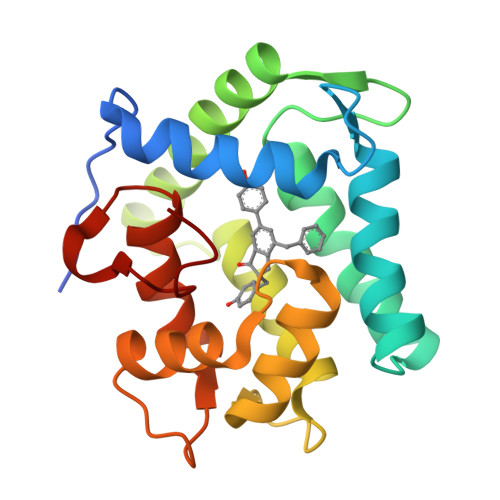
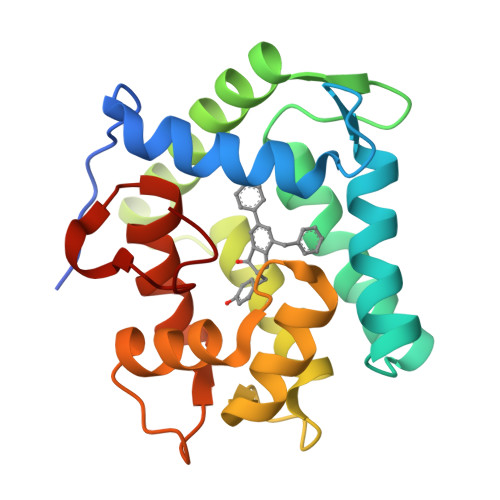
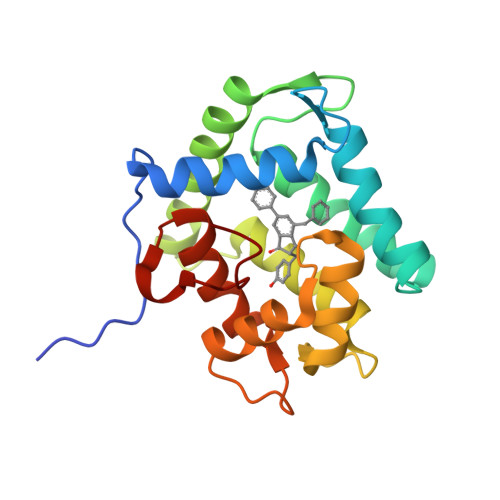
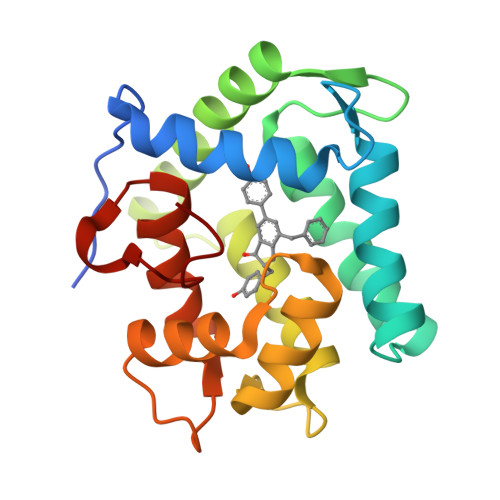
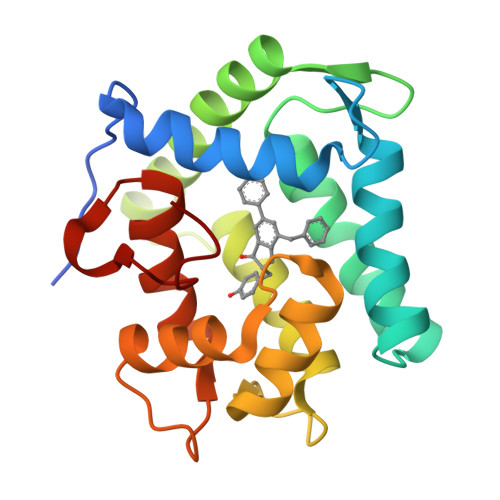
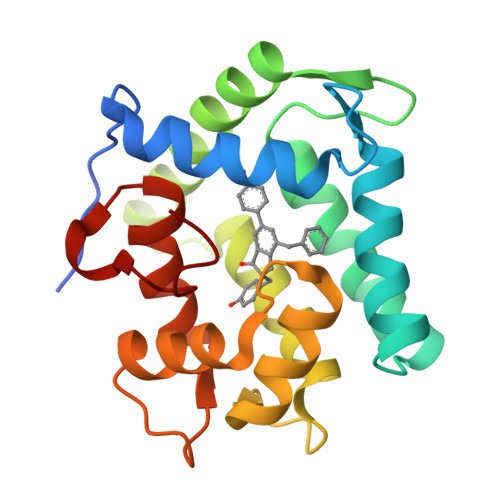
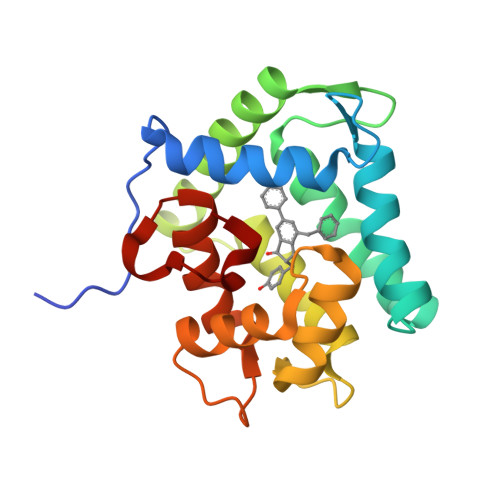
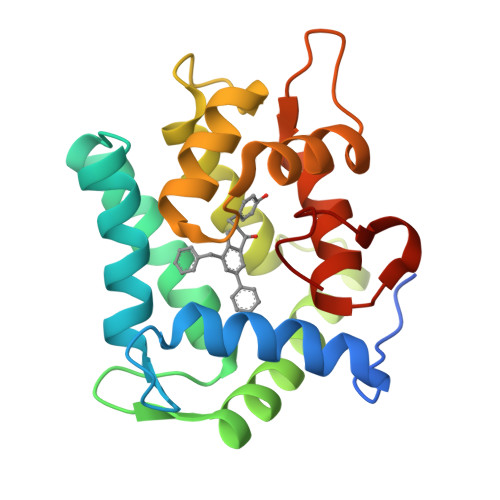
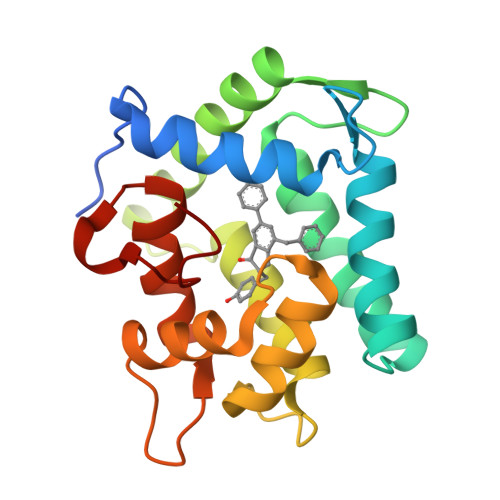
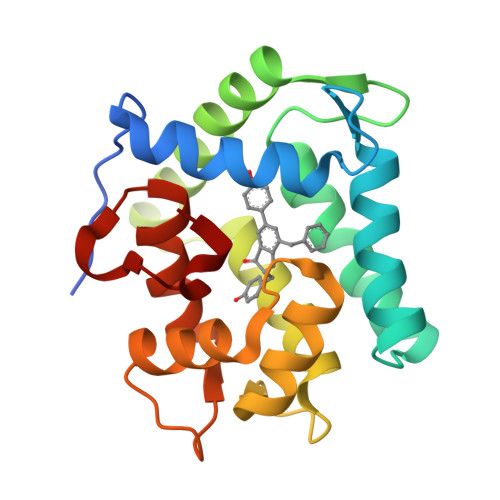
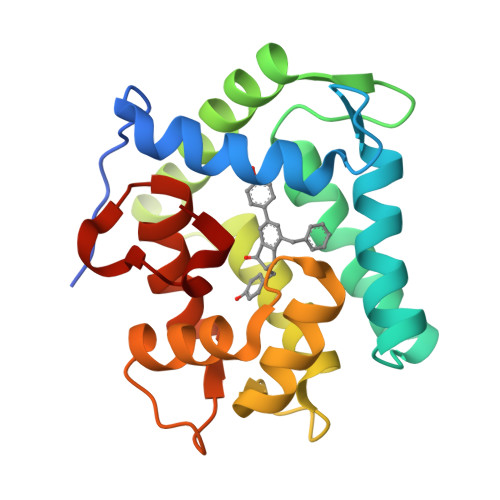
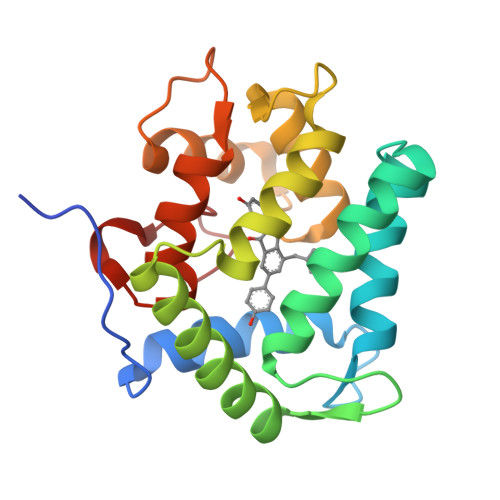
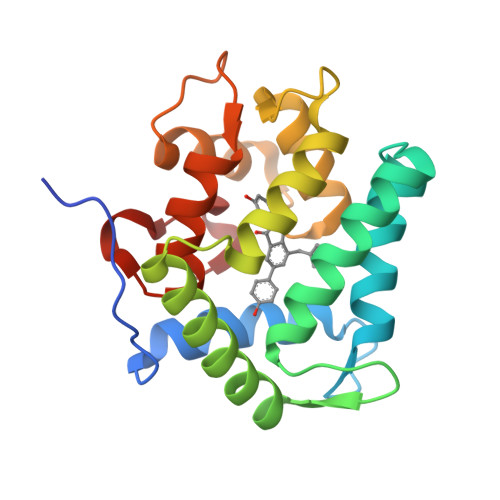
The Ca2+-binding photoprotein aequorin is a complex of apoAequorin (apoprotein) and (S)-2-peroxycoelenterazine. Aequorin can be regenerated by the incubation of apoAequorin with coelenterazine and molecular oxygen (O2). In this study, to investigate the molecular recognition of apoAequorin for coelenterazine using chemical probes, the chiral deaza-analogs of (S)- and (R)-deaza-CTZ (daCTZ) for coelenterazine and of (S)-2- and (R)-2-hydroxymethyl-deaza-CTZ (HM-daCTZ) for 2-peroxycoelenterazine were efficiently prepared by the improvement method. The chiral deaza-analogs of (S)-daCTZ and (S)-HM-daCTZ selectively inhibited the regeneration step to aequorin by binding the catalytic site of coelenterazine in the apoAequorin molecule. The crystal structures of the apoAequorin complexes with (S)-daCTZ and (S)-HM-daCTZ were determined, suggesting that the hydroxy moiety at the C6-hydroxyphenyl group and the carbonyl moiety of the imidazopyrazinone ring in coelenterazine are essential to bind the apoAequorin molecule through hydrogen bonding. Therefore, the chiral deaza-analogs of coelenterazine can be used as a probe to study the interaction between coelenterazine and the related proteins including photoprotein, luciferase, and coelenterazine-binding protein.
Organizational Affiliation:
Yokohama Research Center, JNC Co., Yokohama, Japan.