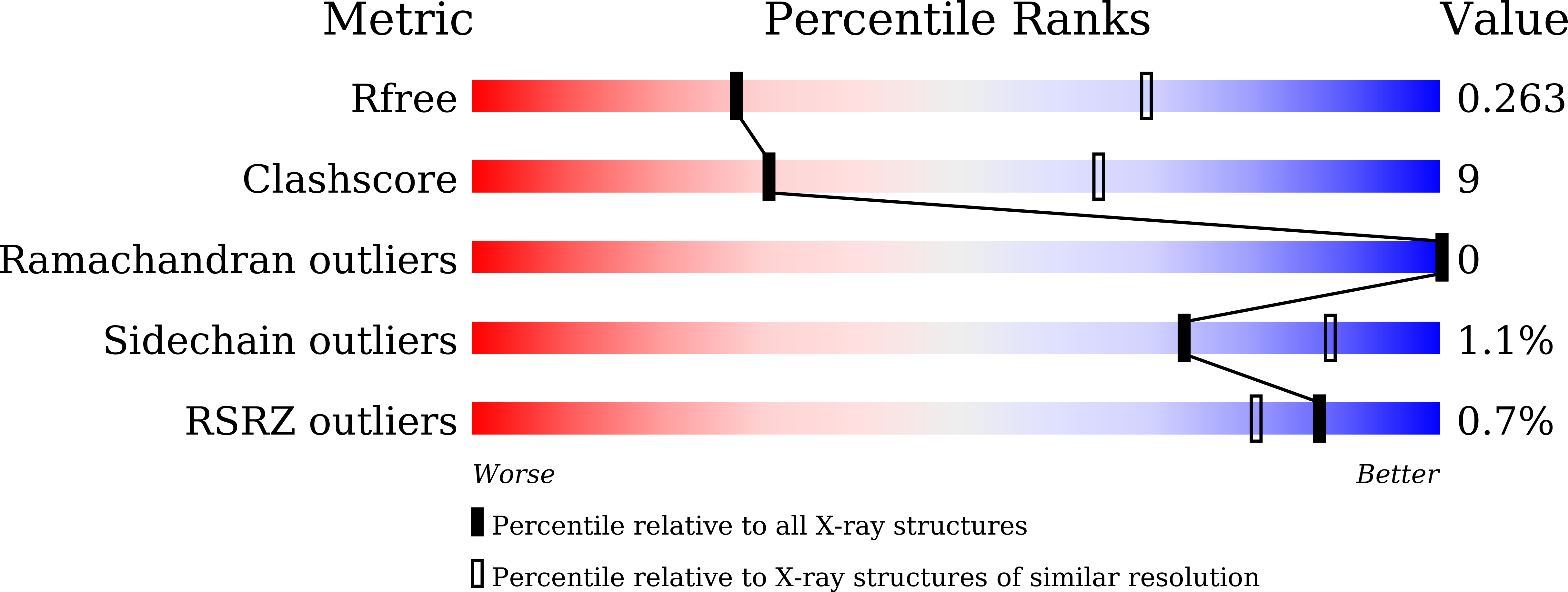
Structural basis for SdgB- and SdgA-mediated glycosylation of staphylococcal adhesive proteins.
Kim, D.G., Baek, I., Lee, Y., Kim, H., Kim, J.Y., Bang, G., Kim, S., Yoon, H.J., Han, B.W., Suh, S.W., Kim, H.S.(2021) Acta Crystallogr D Struct Biol 77: 1460-1474
- PubMed: 34726173
- DOI: https://doi.org/10.1107/S2059798321010068
- Primary Citation of Related Structures:
7EC1, 7EC3, 7EC6, 7EC7, 7VFK, 7VFL, 7VFM, 7VFN, 7VFO - PubMed Abstract:
The initiation of infection of host tissues by Staphylococcus aureus requires a family of staphylococcal adhesive proteins containing serine-aspartate repeat (SDR) domains, such as ClfA. The O-linked glycosylation of the long-chain SDR domain mediated by SdgB and SdgA is a key virulence factor that protects the adhesive SDR proteins against host proteolytic attack in order to promote successful tissue colonization, and has also been implicated in staphylococcal agglutination, which leads to sepsis and an immunodominant epitope for a strong antibody response. Despite the biological significance of these two glycosyltransferases involved in pathogenicity and avoidance of the host innate immune response, their structures and the molecular basis of their activity have not been investigated. This study reports the crystal structures of SdgB and SdgA from S. aureus as well as multiple structures of SdgB in complex with its substrates (for example UDP, N-acetylglucosamine or SDR peptides), products (glycosylated SDR peptides) or phosphate ions. Together with biophysical and biochemical analyses, this structural work uncovered the novel mechanism by which SdgB and SdgA carry out the glycosyl-transfer process to the long SDR region in SDR proteins. SdgB undergoes dynamic changes in its structure such as a transition from an open to a closed conformation upon ligand binding and takes diverse forms, both as a homodimer and as a heterodimer with SdgA. Overall, these findings not only elucidate the putative role of the three domains of SdgB in recognizing donor and acceptor substrates, but also provide new mechanistic insights into glycosylation of the SDR domain, which can serve as a starting point for the development of antibacterial drugs against staphylococcal infections.
Organizational Affiliation:
Research Institute, National Cancer Center, Goyang, Gyeonggi 10408, Republic of Korea.