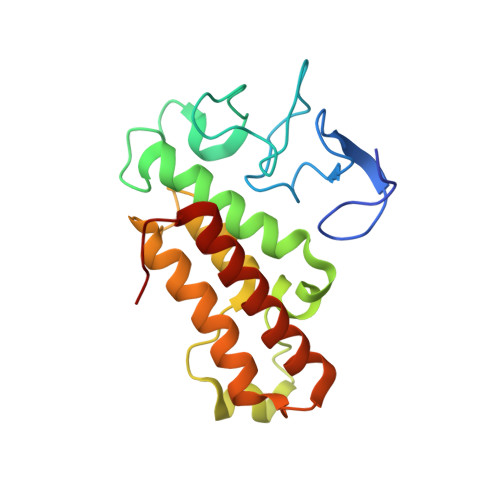
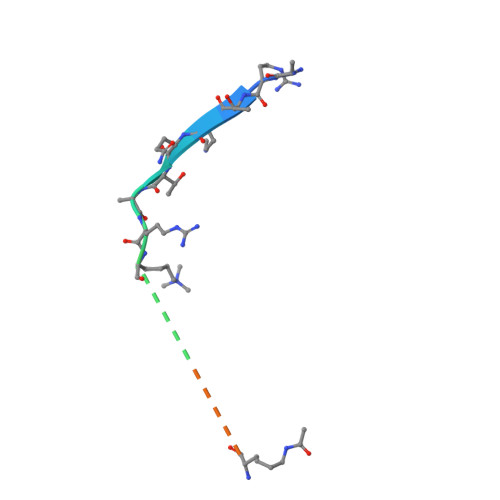
A TRIM66/DAX1/Dux axis suppresses the totipotent 2-cell-like state in murine embryonic stem cells.
Zuo, F., Jiang, J., Fu, H., Yan, K., Liefke, R., Zhang, J., Hong, Y., Chang, Z., Liu, N., Wang, Z., Xi, Q.(2022) Cell Stem Cell 29: 948-961.e6
- PubMed: 35659877
- DOI: https://doi.org/10.1016/j.stem.2022.05.004
- Primary Citation of Related Structures:
7EBJ, 7EBK - PubMed Abstract:
2-cell-like cells (2CLCs)-which comprise only ∼1% of murine embryonic stem cells (mESCs)-resemble blastomeres of 2-cell-stage embryos and are used to investigate zygotic genome activation (ZGA). Here, we discovered that TRIM66 and DAX1 function together as negative regulators of the 2C-like state in mESCs. Chimeric assays confirmed that mESCs lacking TRIM66 or DAX1 function have bidirectional embryonic and extraembryonic differentiation potential. TRIM66 functions by recruiting the co-repressor DAX1 to the Dux promoter, and TRIM66's repressive effect on Dux is dependent on DAX1. A solved crystal structural shows that TRIM66's PHD finger recognizes H3K4-K9me3, and mutational evidence confirmed that TRIM66's PHD finger is essential for its repression of Dux. Thus, beyond expanding the scope of known 2CLC regulators, our study demonstrates that interventions disrupting TRIM66 or DAX1 function in mESCs yield 2CLCs with expanded bidirectional differentiation potential, opening doors for the practical application of these totipotent-like cells.
Organizational Affiliation:
Joint Graduate Program of Peking-Tsinghua-NIBS, Tsinghua University, Beijing 100084, China.