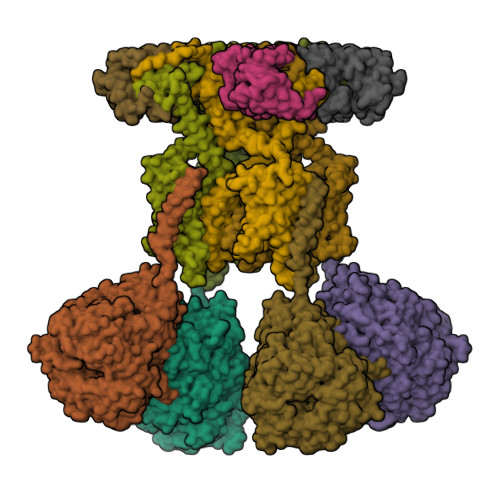
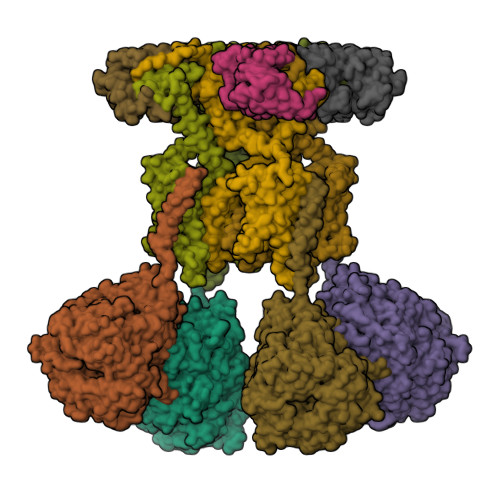
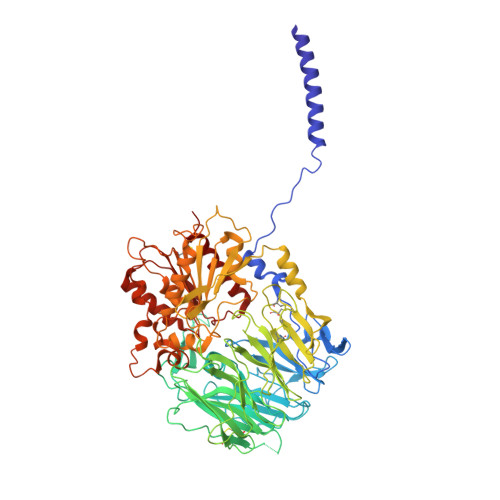
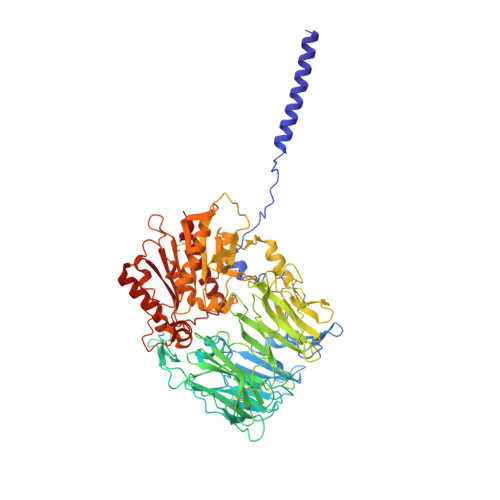
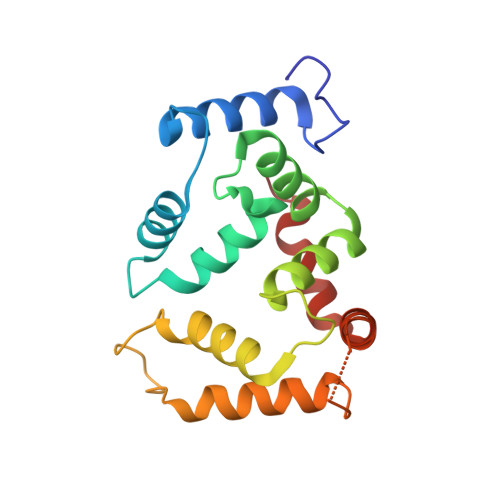
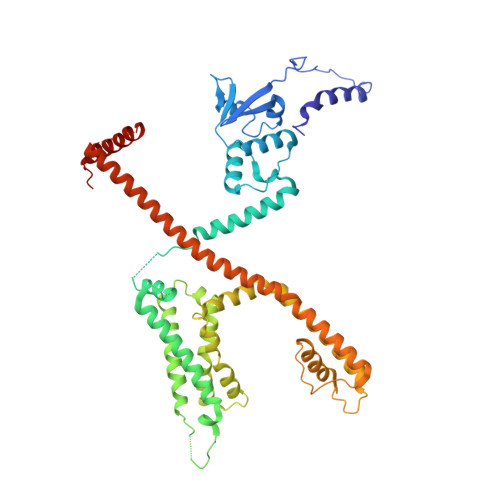
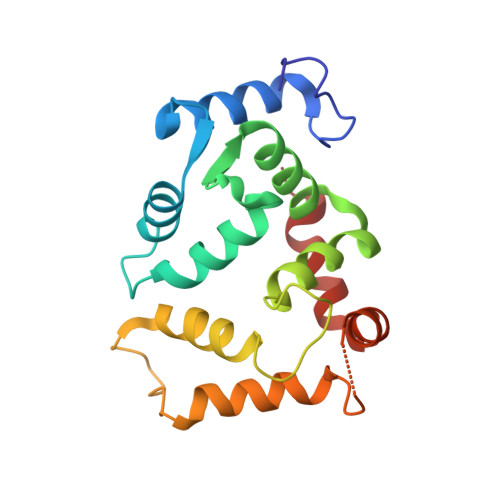
Structural basis of gating modulation of Kv4 channel complexes.
Kise, Y., Kasuya, G., Okamoto, H.H., Yamanouchi, D., Kobayashi, K., Kusakizako, T., Nishizawa, T., Nakajo, K., Nureki, O.(2021) Nature 599: 158-164
- PubMed: 34552243
- DOI: https://doi.org/10.1038/s41586-021-03935-z
- Primary Citation of Related Structures:
7E7Z, 7E83, 7E84, 7E87, 7E89, 7E8B, 7E8E, 7E8G, 7E8H, 7F0J, 7F3F - PubMed Abstract:
Modulation of voltage-gated potassium (Kv) channels by auxiliary subunits is central to the physiological function of channels in the brain and heart 1,2 . Native Kv4 tetrameric channels form macromolecular ternary complexes with two auxiliary β-subunits-intracellular Kv channel-interacting proteins (KChIPs) and transmembrane dipeptidyl peptidase-related proteins (DPPs)-to evoke rapidly activating and inactivating A-type currents, which prevent the backpropagation of action potentials 1-5 . However, the modulatory mechanisms of Kv4 channel complexes remain largely unknown. Here we report cryo-electron microscopy structures of the Kv4.2-DPP6S-KChIP1 dodecamer complex, the Kv4.2-KChIP1 and Kv4.2-DPP6S octamer complexes, and Kv4.2 alone. The structure of the Kv4.2-KChIP1 complex reveals that the intracellular N terminus of Kv4.2 interacts with its C terminus that extends from the S6 gating helix of the neighbouring Kv4.2 subunit. KChIP1 captures both the N and the C terminus of Kv4.2. In consequence, KChIP1 would prevent N-type inactivation and stabilize the S6 conformation to modulate gating of the S6 helices within the tetramer. By contrast, unlike the reported auxiliary subunits of voltage-gated channel complexes, DPP6S interacts with the S1 and S2 helices of the Kv4.2 voltage-sensing domain, which suggests that DPP6S stabilizes the conformation of the S1-S2 helices. DPP6S may therefore accelerate the voltage-dependent movement of the S4 helices. KChIP1 and DPP6S do not directly interact with each other in the Kv4.2-KChIP1-DPP6S ternary complex. Thus, our data suggest that two distinct modes of modulation contribute in an additive manner to evoke A-type currents from the native Kv4 macromolecular complex.
Organizational Affiliation:
Department of Biological Sciences, Graduate School of Science, The University of Tokyo, Tokyo, Japan. yoshiaki.kise@bs.s.u-toyko.ac.jp.