Crystal structures of a nicotine MATE transporter provide insight into its mechanism of substrate transport.
Tanaka, Y., Iwaki, S., Sasaki, A., Tsukazaki, T.(2021) FEBS Lett 595: 1902-1913
- PubMed: 34050946
- DOI: https://doi.org/10.1002/1873-3468.14136
- Primary Citation of Related Structures:
7DQK - PubMed Abstract:
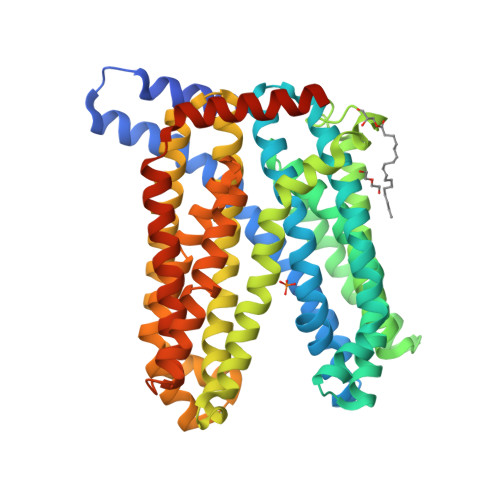
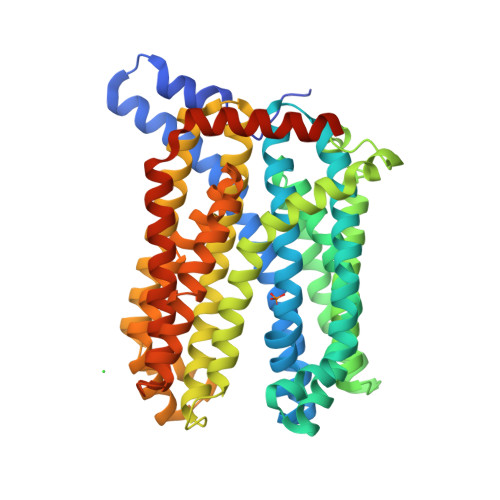
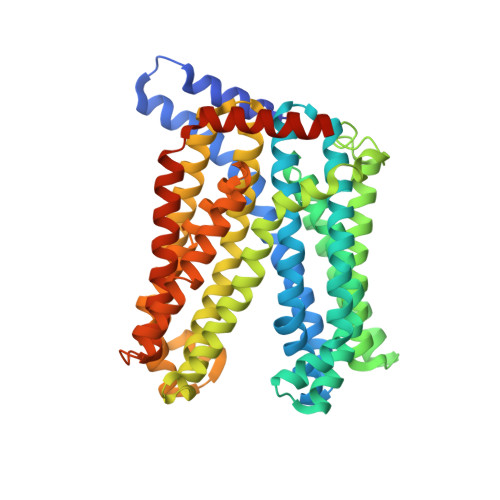
A transporter of the multidrug and toxic compound extrusion (MATE) family, Nicotiana tabacum MATE2 (NtMATE2), is located in the vacuole membrane of the tobacco plant root and is involved in the transportation of nicotine, a secondary or specialized metabolic compound in Solanaceae. Here, we report the crystal structures of NtMATE2 in its outward-facing forms. The overall structure has a bilobate V-shape with pseudo-symmetrical assembly of the N- and C-lobes. In one crystal structure, the C-lobe cavity of NtMATE2 interacts with an unidentified molecule that may partially mimic a substrate. In addition, NtMATE2-specific conformational transitions imply that an unprecedented movement of the transmembrane α-helix 7 is related to the release of the substrate into the vacuolar lumen.
Organizational Affiliation:
Nara Institute of Science and Technology, Ikoma, Japan.