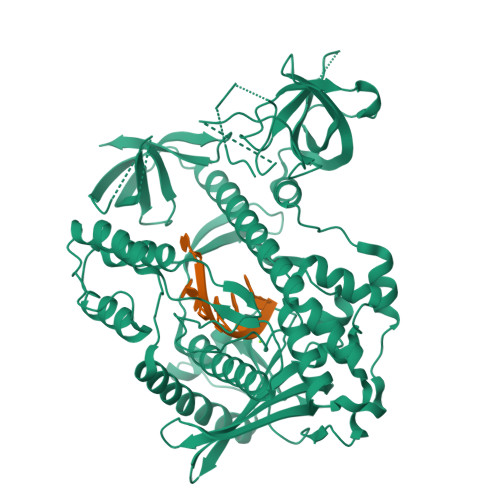
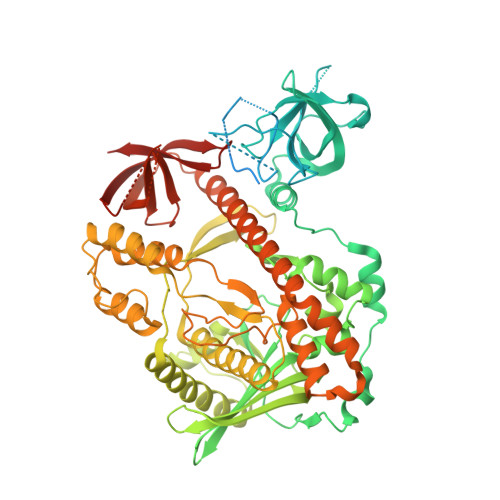
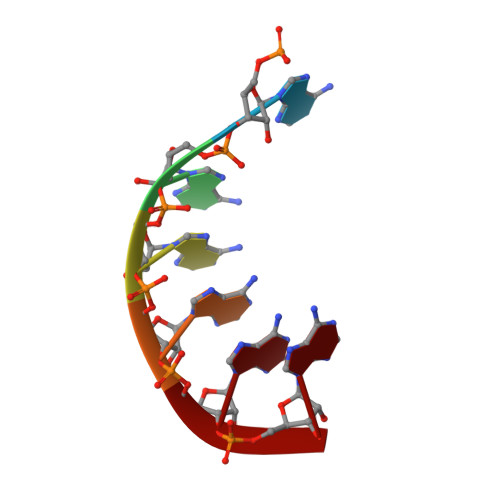
Molecular mechanism of RNase R substrate sensitivity for RNA ribose methylation.
Abula, A., Li, X., Quan, X., Yang, T., Liu, Y., Guo, H., Li, T., Ji, X.(2021) Nucleic Acids Res 49: 4738-4749
- PubMed: 33788943
- DOI: https://doi.org/10.1093/nar/gkab202
- Primary Citation of Related Structures:
7DCY, 7DIC, 7DID, 7DOL - PubMed Abstract:
RNA 2'-O-methylation is widely distributed and plays important roles in various cellular processes. Mycoplasma genitalium RNase R (MgR), a prokaryotic member of the RNase II/RNB family, is a 3'-5' exoribonuclease and is particularly sensitive to RNA 2'-O-methylation. However, how RNase R interacts with various RNA species and exhibits remarkable sensitivity to substrate 2'-O-methyl modifications remains elusive. Here we report high-resolution crystal structures of MgR in apo form and in complex with various RNA substrates. The structural data together with extensive biochemical analysis quantitively illustrate MgR's ribonuclease activity and significant sensitivity to RNA 2'-O-methylation. Comparison to its related homologs reveals an exquisite mechanism for the recognition and degradation of RNA substrates. Through structural and mutagenesis studies, we identified proline 277 to be responsible for the significant sensitivity of MgR to RNA 2'-O-methylation within the RNase II/RNB family. We also generated several MgR variants with modulated activities. Our work provides a mechanistic understanding of MgR activity that can be harnessed as a powerful RNA analytical tool that will open up a new venue for RNA 2'-O-methylations research in biological and clinical samples.
Organizational Affiliation:
The State Key Laboratory of Pharmaceutical Biotechnology, School of Life Sciences, Nanjing University, Nanjing, China.