Structural basis for the exolytic activity of polysaccharide lyase family 6 alginate lyase BcAlyPL6 from human gut microbe Bacteroides clarus.
Wang, B., Dong, S., Li, F.L., Ma, X.Q.(2021) Biochem Biophys Res Commun 547: 111-117
- PubMed: 33610038
- DOI: https://doi.org/10.1016/j.bbrc.2021.02.040
- Primary Citation of Related Structures:
7DMK - PubMed Abstract:
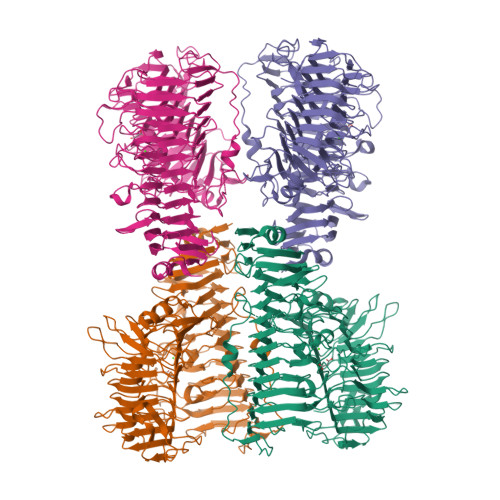
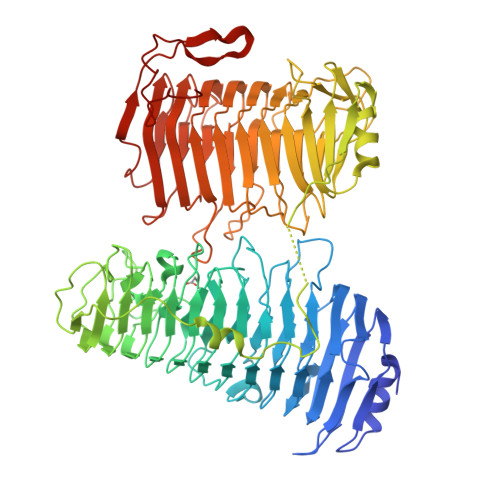
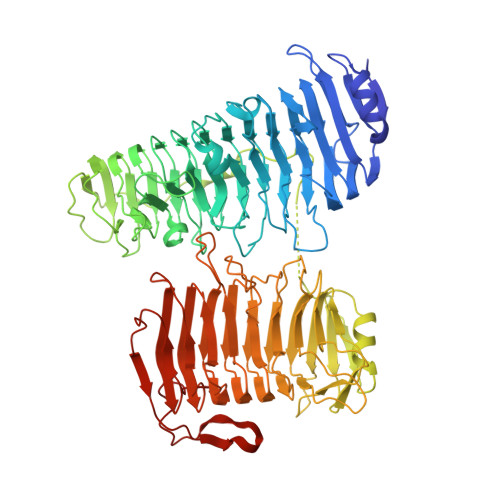
Alginate is the structural polysaccharide of the cell wall of brown algae, which is an important carbon source for marine life. The depolymerization of alginate is dependent on alginate lyases. Recent studies showed that the alginate utilization ability had been obtained by human gut microbes. In contrast to the great number of studies on alginate lyases from marine/soil organisms, studies on alginate lyases from gut microbes are still limited. Here, the structure of a polysaccharide lyase family 6 (PL6) alginate lyase from human gut microbe Bacteroides clarus was solved by X-ray crystallography, which represents the cluster of two-domain PL6 alginate lyases from Bacteroidetes. Similar with the two-domain alginate lyase AlyGC originated from marine bacterium, both the N terminal domain (NTD) and C terminal domain (CTD) of BcAlyPL6 show right-handed parallel β-helix fold. However, unlike AlyGC, which forms a homodimer, BcAlyPL6 functions as a monomer. Biochemical analysis indicates that the substrate binding affinity is mainly contributed by the NTD while the CTD of BcAlyPL6 is involved in the formation of -1 subsite, which is essential for substrate turnover rate. Furthermore, CTD is involved in shaping a closed catalytic pocket, and deletion of it leads to increased activity towards highly polymerized substrate. Structure comparison of PL6 family alginate lyases implies that the linkers of two-domain alginate lyases might have evolutionary relationship with the N/C terminal extension of single-domain lyases.
Organizational Affiliation:
Shandong Provincial Key Laboratory of Energy Genetics, Key Laboratory of Biofuels, Qingdao Institute of Bioenergy and Bioprocess Technology, Chinese Academy of Sciences, Qingdao, 266101, PR China.