Selective inhibition reveals the regulatory function of DYRK2 in protein synthesis and calcium entry.
Wei, T., Wang, J., Liang, R., Chen, W., Chen, Y., Ma, M., He, A., Du, Y., Zhou, W., Zhang, Z., Zeng, X., Wang, C., Lu, J., Guo, X., Chen, X.W., Wang, Y., Tian, R., Xiao, J., Lei, X.(2022) Elife 11
- PubMed: 35439114
- DOI: https://doi.org/10.7554/eLife.77696
- Primary Citation of Related Structures:
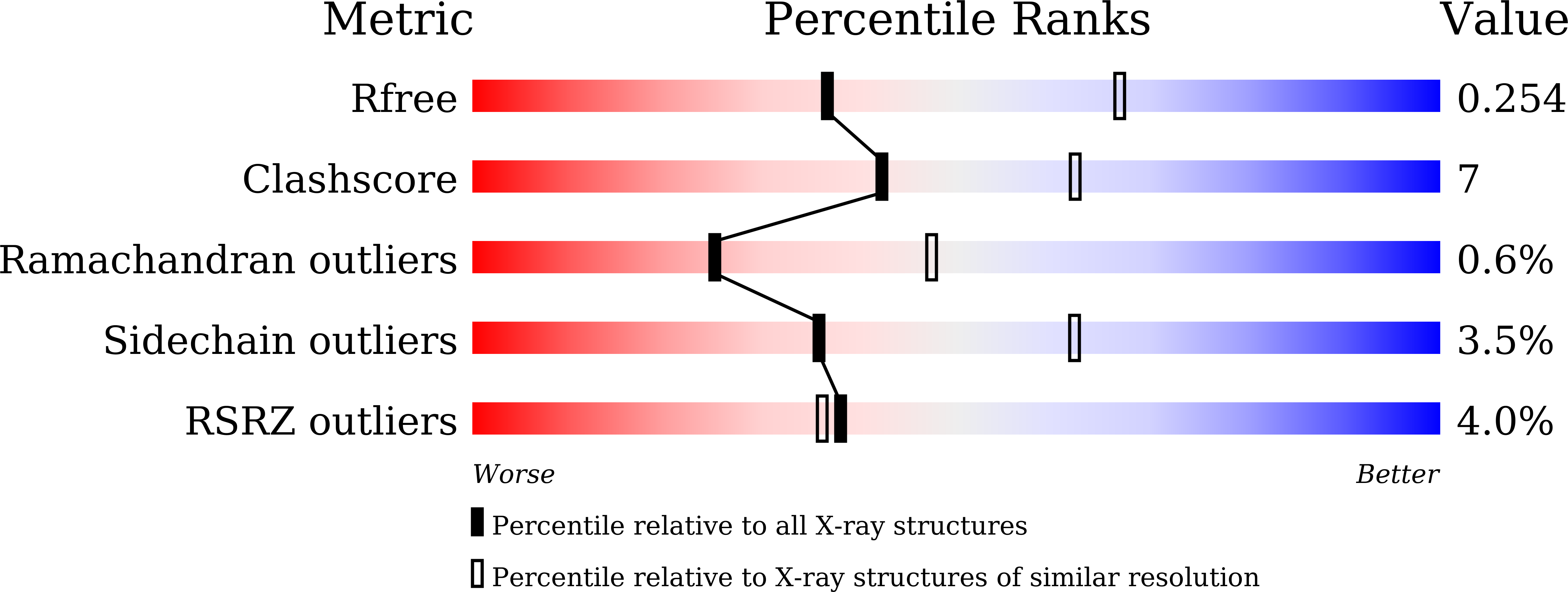
7DG4, 7DH9, 7DHC, 7DHH, 7DHK, 7DHN, 7DHO, 7DHV, 7DJO, 7DL6 - PubMed Abstract:
The dual-specificity tyrosine phosphorylation-regulated kinase DYRK2 has emerged as a critical regulator of cellular processes. We took a chemical biology approach to gain further insights into its function. We developed C17, a potent small-molecule DYRK2 inhibitor, through multiple rounds of structure-based optimization guided by several co-crystallized structures. C17 displayed an effect on DYRK2 at a single-digit nanomolar IC 50 and showed outstanding selectivity for the human kinome containing 467 other human kinases. Using C17 as a chemical probe, we further performed quantitative phosphoproteomic assays and identified several novel DYRK2 targets, including eukaryotic translation initiation factor 4E-binding protein 1 (4E-BP1) and stromal interaction molecule 1 (STIM1). DYRK2 phosphorylated 4E-BP1 at multiple sites, and the combined treatment of C17 with AKT and MEK inhibitors showed synergistic 4E-BP1 phosphorylation suppression. The phosphorylation of STIM1 by DYRK2 substantially increased the interaction of STIM1 with the ORAI1 channel, and C17 impeded the store-operated calcium entry process. These studies collectively further expand our understanding of DYRK2 and provide a valuable tool to pinpoint its biological function.
Organizational Affiliation:
The State Key Laboratory of Protein and Plant Gene Research, School of Life Sciences, Peking University, Beijing, China.