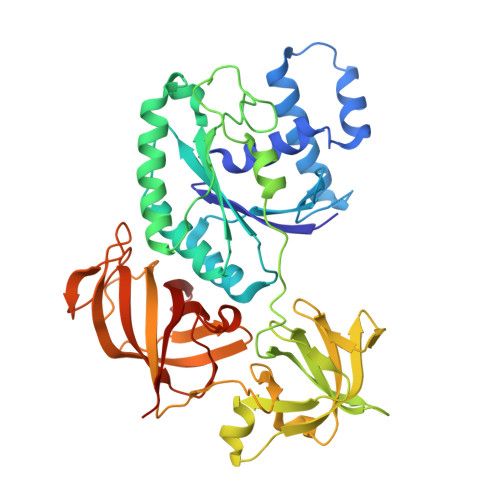
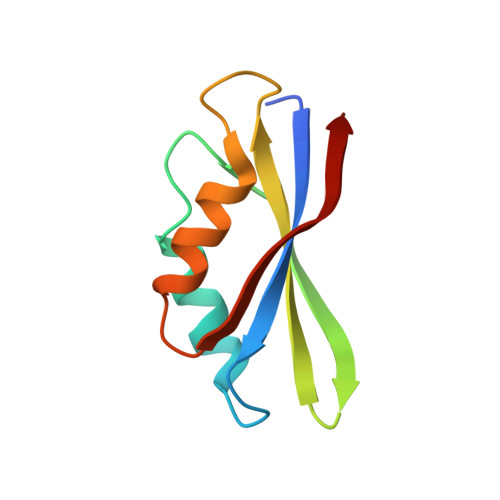
Structural insights into the Switching Off of the Interaction between the Archaeal Ribosomal Stalk and aEF1A by Nucleotide Exchange Factor aEF1B.
Suzuki, T., Ito, K., Miyoshi, T., Murakami, R., Uchiumi, T.(2021) J Mol Biol 433: 167046-167046
- PubMed: 33971210
- DOI: https://doi.org/10.1016/j.jmb.2021.167046
- Primary Citation of Related Structures:
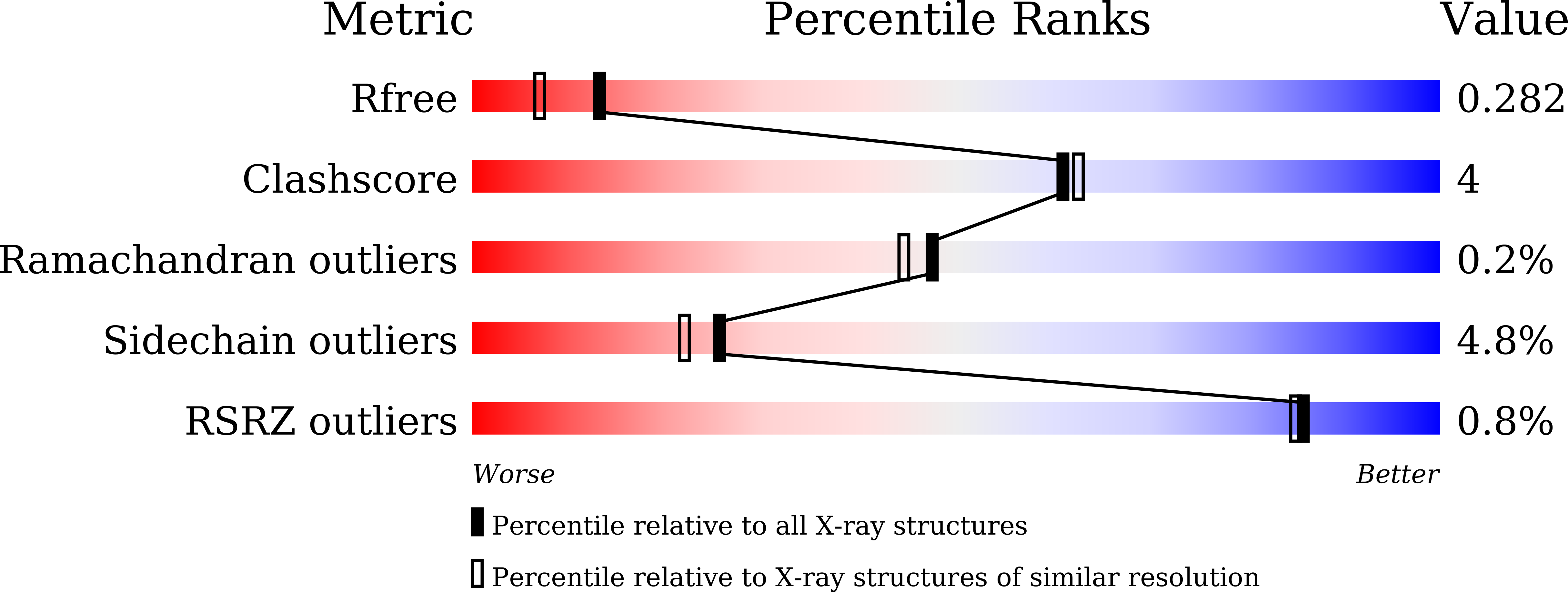
7CSL - PubMed Abstract:
The ribosomal stalk protein plays a crucial role in functional interactions with translational GTPase factors. It has been shown that the archaeal stalk aP1 binds to both GDP- and GTP-bound conformations of aEF1A through its C-terminal region in two different modes. To obtain an insight into how the aP1•aEF1A binding mode changes during the process of nucleotide exchange from GDP to GTP on aEF1A, we have analyzed structural changes in aEF1A upon binding of the nucleotide exchange factor aEF1B. The isolated archaeal aEF1B has nucleotide exchange ability in the presence of aa-tRNA but not deacylated tRNA, and increases activity of polyphenylalanine synthesis 4-fold. The aEF1B mutation, R90A, results in loss of its original nucleotide exchange activity but retains a remarkable ability to enhance polyphenylalanine synthesis. These results suggest an additional functional role for aEF1B other than in nucleotide exchange. The crystal structure of the aEF1A•aEF1B complex, resolved at 2.0 Å resolution, shows marked rotational movement of domain 1 of aEF1A compared to the structure of aEF1A•GDP•aP1, and this conformational change results in disruption of the original aP1 binding site between domains 1 and 3 of aEF1A. The loss of aP1 binding to the aEF1A•aEF1B complex was confirmed by native gel analysis. The results suggest that aEF1B plays a role in switching off the interaction between aP1 and aEF1A•GDP, as well as in nucleotide exchange, and promote translation elongation.
Organizational Affiliation:
Department of Biology, Faculty of Science, Niigata University, Ikarashi 2-8050, Nishi-ku, Niigata 950-2181, Japan.