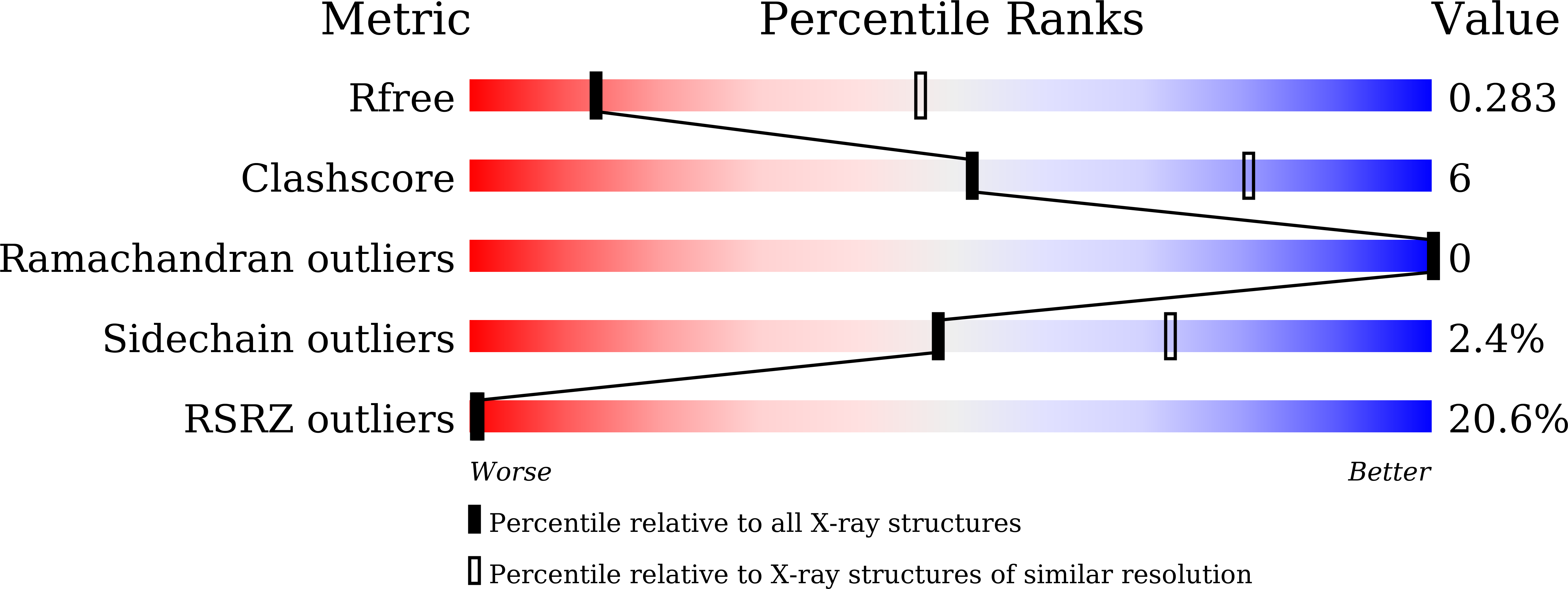
Structural basis for transcriptional coactivator recognition by SMAD2 in TGF-beta signaling.
Miyazono, K.I., Ito, T., Fukatsu, Y., Wada, H., Kurisaki, A., Tanokura, M.(2020) Sci Signal 13
- PubMed: 33323411
- DOI: https://doi.org/10.1126/scisignal.abb9043
- Primary Citation of Related Structures:
6M64, 7CO1 - PubMed Abstract:
Transforming growth factor-β (TGF-β) proteins regulate multiple cellular functions, including cell proliferation, apoptosis, and extracellular matrix formation. The dysregulation of TGF-β signaling causes diseases such as cancer and fibrosis, and therefore, understanding the biochemical basis of TGF-β signal transduction is important for elucidating pathogenic mechanisms in these diseases. SMAD proteins are transcription factors that mediate TGF-β signaling-dependent gene expression. The transcriptional coactivator CBP directly interacts with the MH2 domains of SMAD2 to activate SMAD complex-dependent gene expression. Here, we report the structural basis for CBP recognition by SMAD2. The crystal structures of the SMAD2 MH2 domain in complex with the SMAD2-binding region of CBP showed that CBP forms an amphiphilic helix on the hydrophobic surface of SMAD2. The expression of a mutated CBP peptide that showed increased SMAD2 binding repressed SMAD2-dependent gene expression in response to TGF-β signaling in cultured cells. Disrupting the interaction between SMAD2 and CBP may therefore be a promising strategy for suppressing SMAD-dependent gene expression.
Organizational Affiliation:
Department of Applied Biological Chemistry, Graduate School of Agricultural and Life Sciences, The University of Tokyo, Tokyo 113-8657, Japan.