Structural basis of the protochromic green/red photocycle of the chromatic acclimation sensor RcaE.
Nagae, T., Unno, M., Koizumi, T., Miyanoiri, Y., Fujisawa, T., Masui, K., Kamo, T., Wada, K., Eki, T., Ito, Y., Hirose, Y., Mishima, M.(2021) Proc Natl Acad Sci U S A 118
- PubMed: 33972439
- DOI: https://doi.org/10.1073/pnas.2024583118
- Primary Citation of Related Structures:
7CKV - PubMed Abstract:
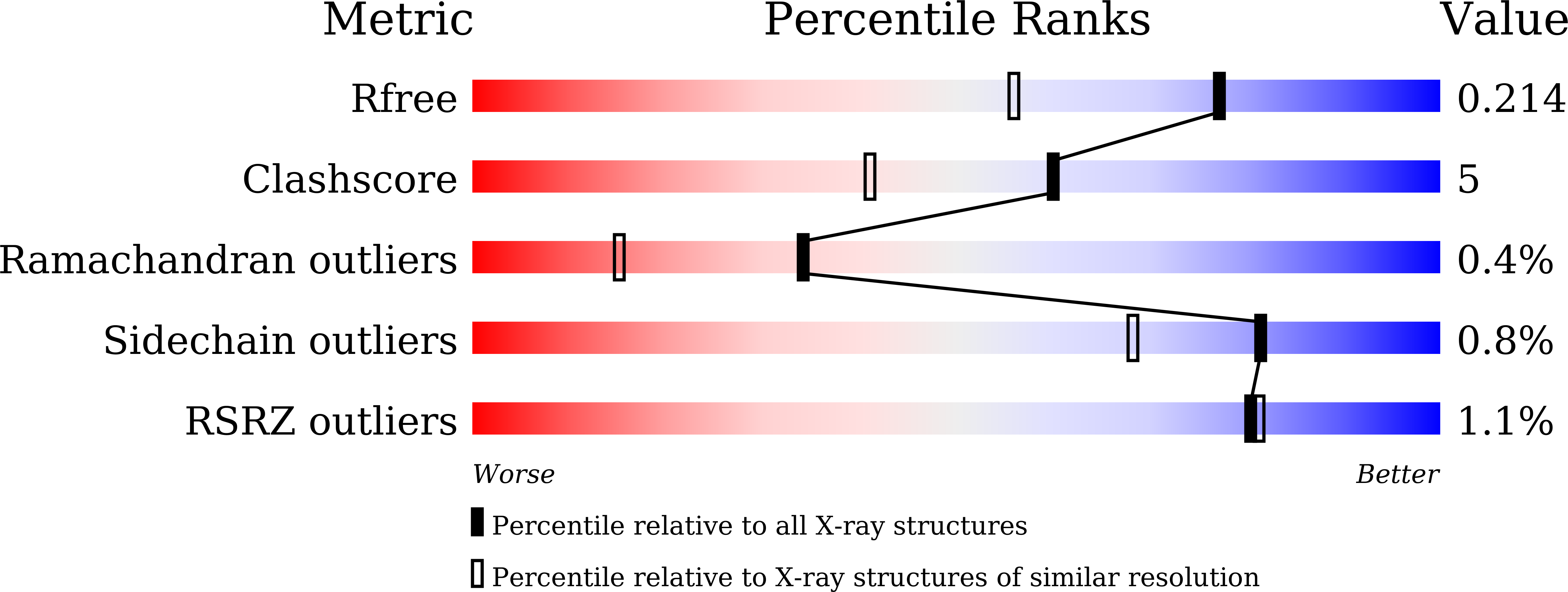
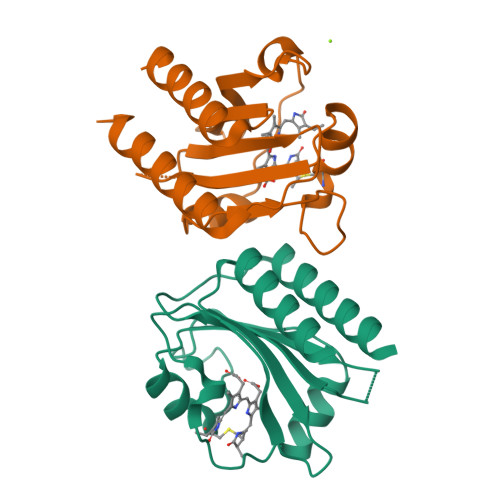
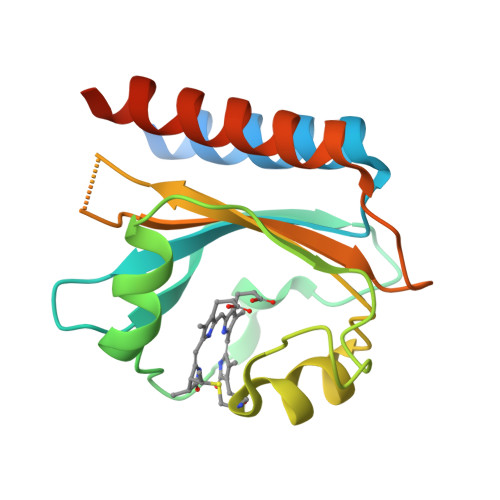
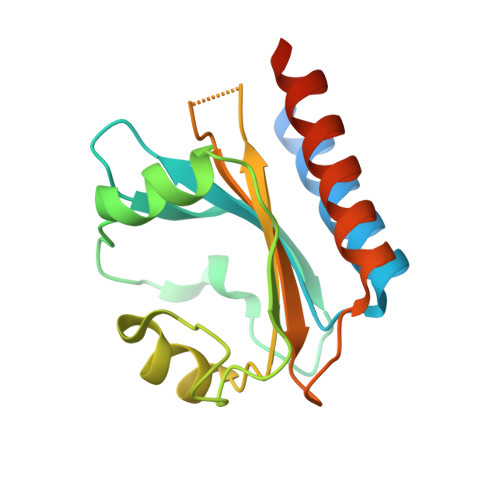
Cyanobacteriochromes (CBCRs) are bilin-binding photosensors of the phytochrome superfamily that show remarkable spectral diversity. The green/red CBCR subfamily is important for regulating chromatic acclimation of photosynthetic antenna in cyanobacteria and is applied for optogenetic control of gene expression in synthetic biology. It is suggested that the absorption change of this subfamily is caused by the bilin C15 -Z /C15- E photoisomerization and a subsequent change in the bilin protonation state. However, structural information and direct evidence of the bilin protonation state are lacking. Here, we report a high-resolution (1.63Å) crystal structure of the bilin-binding domain of the chromatic acclimation sensor RcaE in the red-absorbing photoproduct state. The bilin is buried within a "bucket" consisting of hydrophobic residues, in which the bilin configuration/conformation is C5- Z , syn /C10- Z , syn /C15- E , syn with the A- through C-rings coplanar and the D-ring tilted. Three pyrrole nitrogens of the A- through C-rings are covered in the α-face with a hydrophobic lid of Leu249 influencing the bilin p K a , whereas they are directly hydrogen bonded in the β-face with the carboxyl group of Glu217. Glu217 is further connected to a cluster of waters forming a hole in the bucket, which are in exchange with solvent waters in molecular dynamics simulation. We propose that the "leaky bucket" structure functions as a proton exit/influx pathway upon photoconversion. NMR analysis demonstrated that the four pyrrole nitrogen atoms are indeed fully protonated in the red-absorbing state, but one of them, most likely the B-ring nitrogen, is deprotonated in the green-absorbing state. These findings deepen our understanding of the diverse spectral tuning mechanisms present in CBCRs.
Organizational Affiliation:
Synchrotron Radiation Research Center, Nagoya University, Nagoya 464-8603, Japan.