Structural basis of tetanus toxin neutralization by native human monoclonal antibodies.
Wang, Y., Wu, C., Yu, J., Lin, S., Liu, T., Zan, L., Li, N., Hong, P., Wang, X., Jia, Z., Li, J., Wang, Y., Zhang, M., Yuan, X., Li, C., Xu, W., Zheng, W., Wang, X., Liao, H.X.(2021) Cell Rep 35: 109070-109070
- PubMed: 33951441
- DOI: https://doi.org/10.1016/j.celrep.2021.109070
- Primary Citation of Related Structures:
7CE2 - PubMed Abstract:
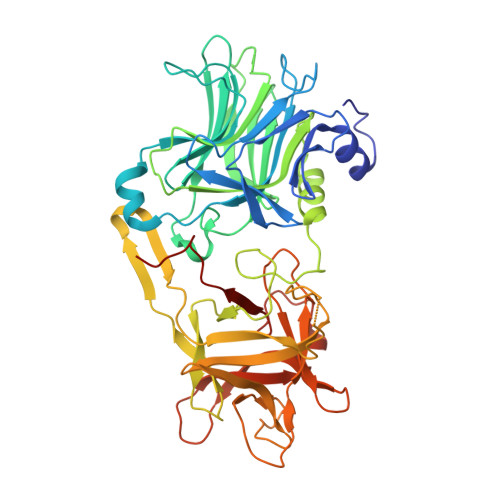
Four potent native human monoclonal antibodies (mAbs) targeting distinct epitopes on tetanus toxin (TeNT) are isolated with neutralization potency ranging from approximately 17 mg to 6 mg each that are equivalent to 250 IU of human anti-TeNT immunoglobulin. TT0170 binds fragment B, and TT0069 and TT0155 bind fragment AB. mAb TT0067 binds fragment C and blocks the binding of TeNT to gangliosides. The co-crystal structure of TT0067 with fragment C of TeNT at a 2.0-Å resolution demonstrates that mAb TT0067 directly occupies the W pocket of one of the receptor binding sites on TeNT, resulting in blocking the binding of TeNT to ganglioside on the surface of host cells. This study reveals at the atomic level the mechanism of action by the TeNT neutralizing antibody. The key neutralization epitope on the fragment C of TeNT identified in our work provides the critical information for the development of fragment C of TeNT as a better and safer tetanus vaccine.
Organizational Affiliation:
Department of Cell Biology, College of Life Science and Technology, Jinan University, Guangzhou 510632, China; Trinomab Biotech Co., Ltd, Zhuhai 519040, China.