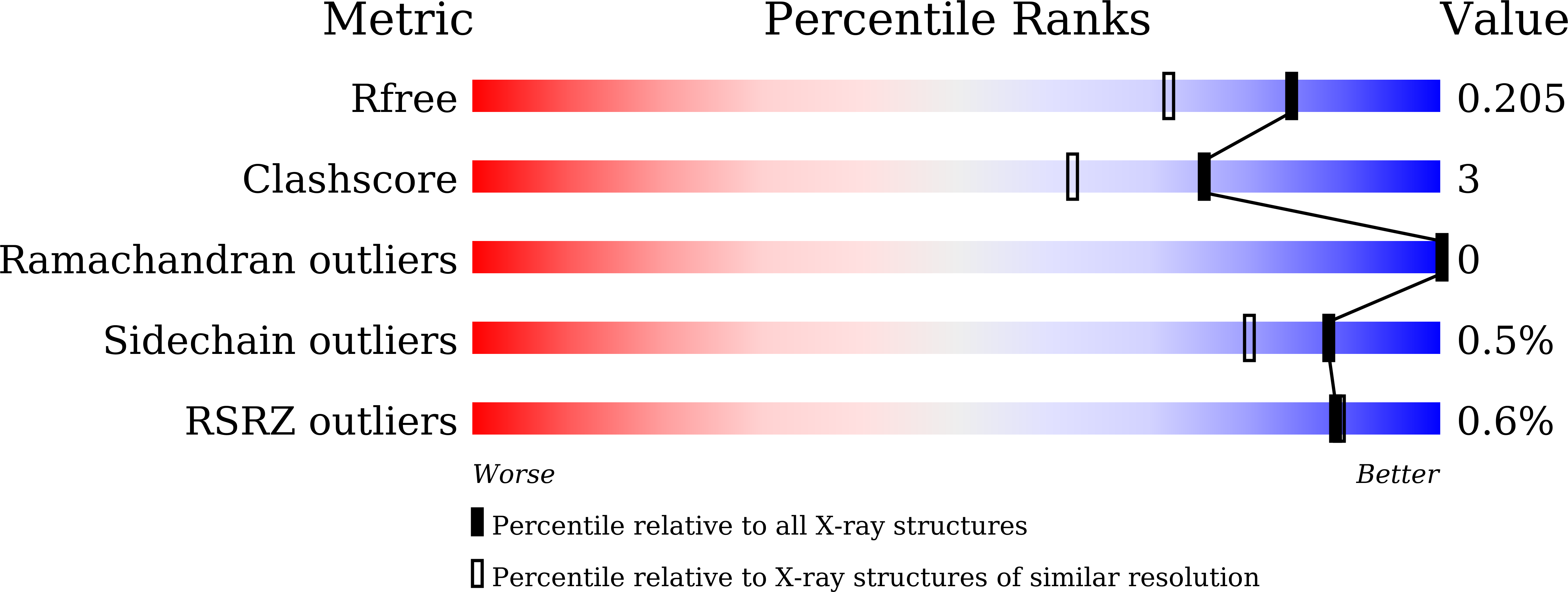
Conformational Trapping of a beta-Glucosides-Binding Protein Unveils the Selective Two-Step Ligand-Binding Mechanism of ABC Importers.
Chandravanshi, M., Samanta, R., Kanaujia, S.P.(2020) J Mol Biol 432: 5711-5734
- PubMed: 32866452
- DOI: https://doi.org/10.1016/j.jmb.2020.08.021
- Primary Citation of Related Structures:
7C63, 7C64, 7C66, 7C67, 7C68, 7C69, 7C6F, 7C6G, 7C6H, 7C6I, 7C6J, 7C6K, 7C6L, 7C6M, 7C6N, 7C6R, 7C6T, 7C6V, 7C6W, 7C6X, 7C6Y, 7C6Z, 7C70, 7C71 - PubMed Abstract:
Substrate-binding proteins (SBPs), selectively capture ligand(s) and ensure their translocation via its cognate ATP-binding cassette (ABC) import system. SBPs bind their cognate ligand(s) via an induced-fit mechanism known as the "Venus Fly-trap"; however, this mechanism lacks the atomic details of all conformational landscape as the confirmatory evidence(s) in its support. In this study, we delineate the atomic details of an SBP, β-glucosides-binding protein (βGlyBP) from Thermus thermophilus HB8. The protein βGlyBP is multi-specific and binds to different types of β-glucosides varying in their glycosidic linkages viz. β-1,2; β-1,3; β-1,4 and β-1,6 with a degree of polymerization of 2-5 glucosyl units. Structurally, the protein βGlyBP possesses four subdomains (N1, N2, C1 and C2). The unliganded protein βGlyBP remains in an open state, which closes upon binding to sophorose (SOP2), laminari-oligosaccharides (LAMn), cello-oligosaccharides (CELn), and gentiobiose (GEN2). This study reports, for the first time, four different structural states (open-unliganded, partial-open-unliganded, open-liganded and closed-liganded) of the protein βGlyBP, revealing its conformational changes upon ligand binding and suggesting a two-step induced-fit mechanism. Further, results suggest that the conformational changes of N1 and C1 subdomains drive the ligand binding, unlike that of the whole N- and C-terminal domains (NTD and CTD) as known in the "Venus Fly-trap" mechanism. Additionally, profiling of stereo-selection mechanism for α- and β-glucosides reveals that in the ligand-binding site four secondary structural elements (L1, H1, H2 and H3) drive the ligand selection. In summary, results demonstrate that the details of conformational changes and ligand selection are pre-encoded in the SBPs.
Organizational Affiliation:
Department of Biosciences and Bioengineering, Indian Institute of Technology Guwahati, Guwahati 781039, Assam, India.