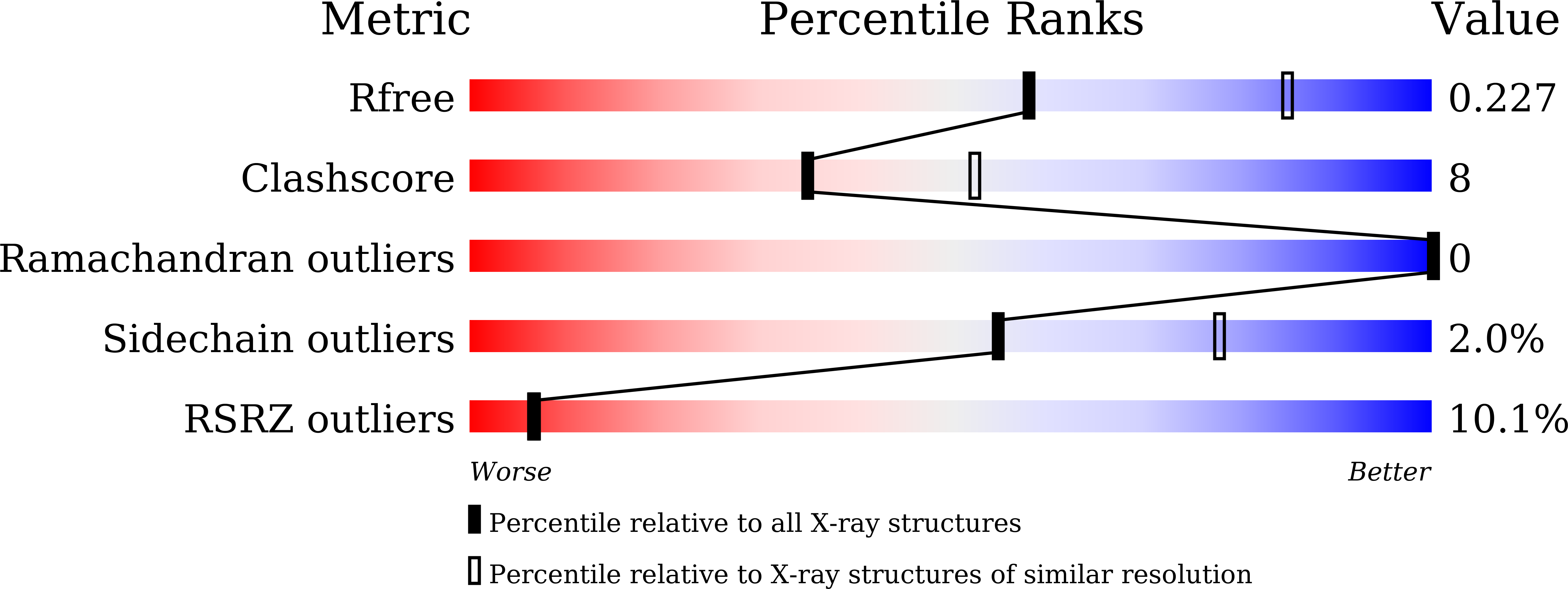
A H-REV107 Peptide Inhibits Tumor Growth and Interacts Directly with Oncogenic KRAS Mutants.
Han, C.W., Jeong, M.S., Ha, S.C., Jang, S.B.(2020) Cancers (Basel) 12
- PubMed: 32486141
- DOI: https://doi.org/10.3390/cancers12061412
- Primary Citation of Related Structures:
7C3Z, 7C40, 7C41 - PubMed Abstract:
Kirsten-RAS (KRAS) has been the target of drugs because it is the most mutated gene in human cancers. Because of the low affinity of drugs for KRAS mutations, it was difficult to target these tumor genes directly. We found a direct interaction between KRAS G12V and tumor suppressor novel H-REV107 peptide with high binding affinity. We report the first crystal structure of an oncogenic mutant, KRAS G12V-H-REV107. This peptide was shown to interact with KRAS G12V in the guanosine diphosphate (GDP)-bound inactive state and to form a stable complex, blocking the activation function of KRAS. We showed that the peptide acted as an inhibitor of mutant KRAS targets by [α- 32 P] guanosine triphosphate (GTP) binding assay. The H-REV107 peptide inhibited pancreatic cancer and colon cancer cell lines in cell proliferation assay. Specially, the H-REV107 peptide can suppress pancreatic tumor growth by reduction of tumor volume and weight in xenotransplantation mouse models. Overall, the results presented herein will facilitate development of novel drugs for inhibition of KRAS mutations in cancer patients.
Organizational Affiliation:
Department of Molecular Biology, College of Natural Sciences, Pusan National University, 2, Busandaehak-ro 63 beon-gil, Geumjeong-gu, Busan 46241, Korea.