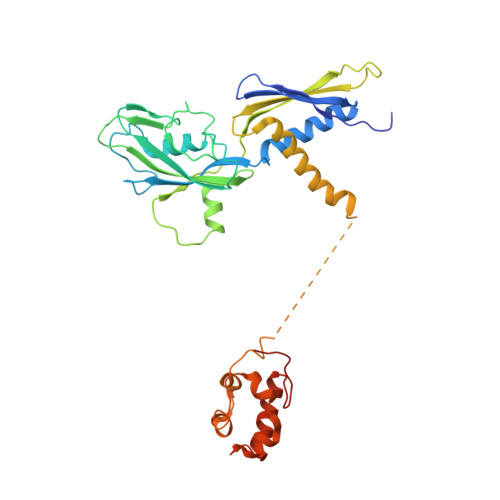
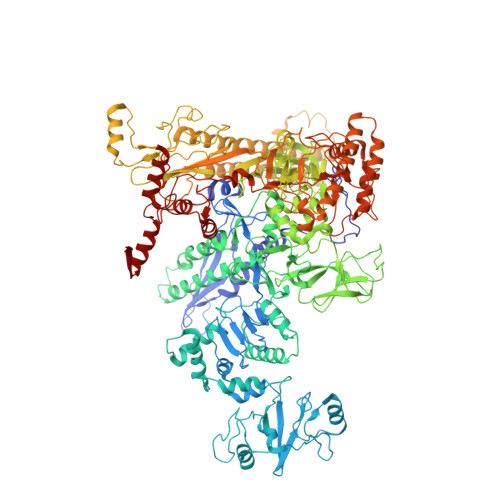
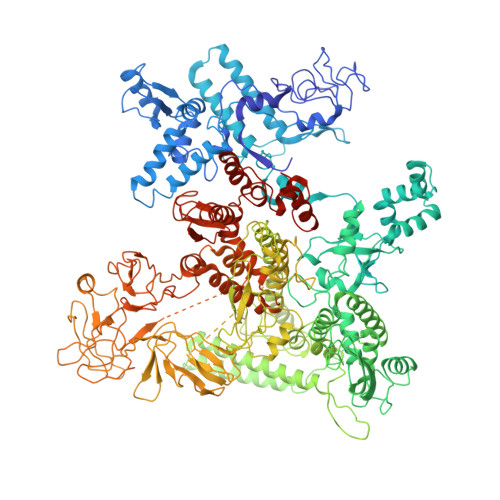
CueR activates transcription through a DNA distortion mechanism.
Fang, C., Philips, S.J., Wu, X., Chen, K., Shi, J., Shen, L., Xu, J., Feng, Y., O'Halloran, T.V., Zhang, Y.(2021) Nat Chem Biol 17: 57-64
- PubMed: 32989300
- DOI: https://doi.org/10.1038/s41589-020-00653-x
- Primary Citation of Related Structures:
6LDI, 7C17 - PubMed Abstract:
The MerR-family transcription factors (TFs) are a large group of bacterial proteins responding to cellular metal ions and multiple antibiotics by binding within central RNA polymerase-binding regions of a promoter. While most TFs alter transcription through protein-protein interactions, MerR TFs are capable of reshaping promoter DNA. To address the question of which mechanism prevails, we determined two cryo-EM structures of transcription activation complexes (TAC) comprising Escherichia coli CueR (a prototype MerR TF), RNAP holoenzyme and promoter DNA. The structures reveal that this TF promotes productive promoter-polymerase association without canonical protein-protein contacts seen between other activator proteins and RNAP. Instead, CueR realigns the key promoter elements in the transcription activation complex by clamp-like protein-DNA interactions: these induce four distinct kinks that ultimately position the -10 element for formation of the transcription bubble. These structural and biochemical results provide strong support for the DNA distortion paradigm of allosteric transcriptional control by MerR TFs.
Organizational Affiliation:
Key Laboratory of Synthetic Biology, CAS Center for Excellence in Molecular Plant Sciences, Shanghai Institute of Plant Physiology and Ecology, Chinese Academy of Sciences, Shanghai, China.