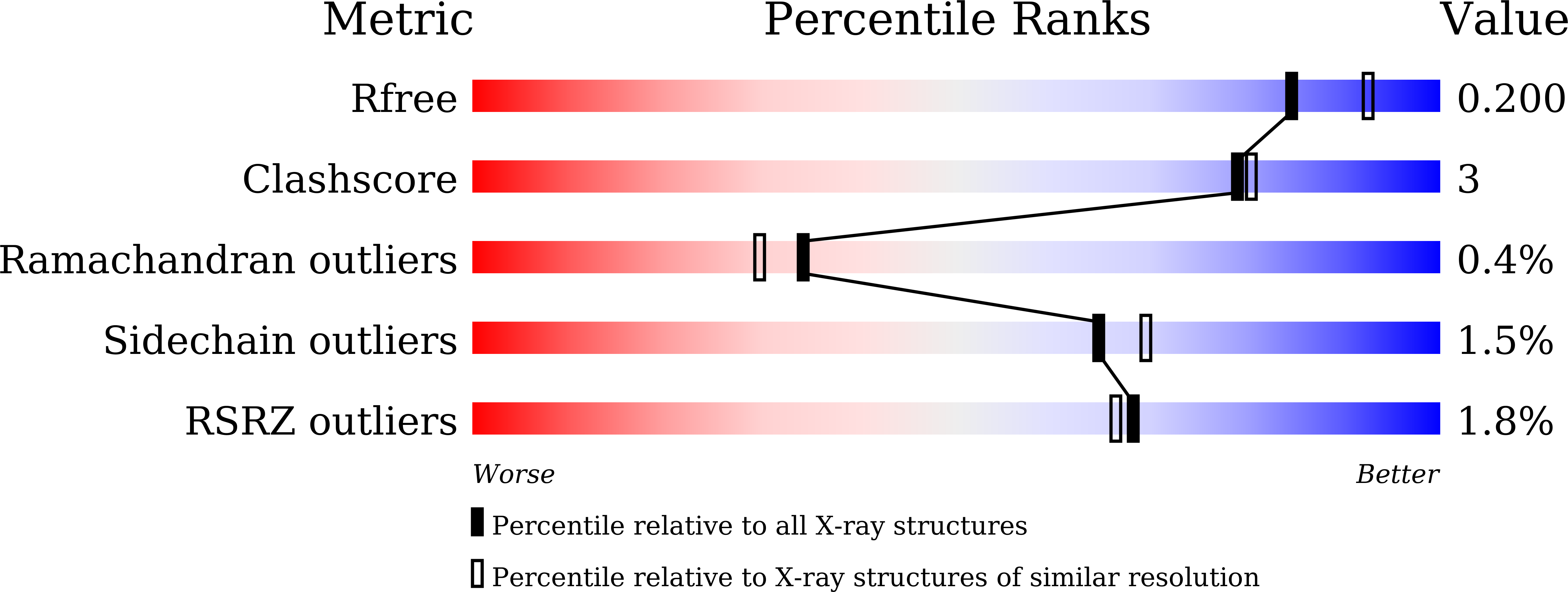
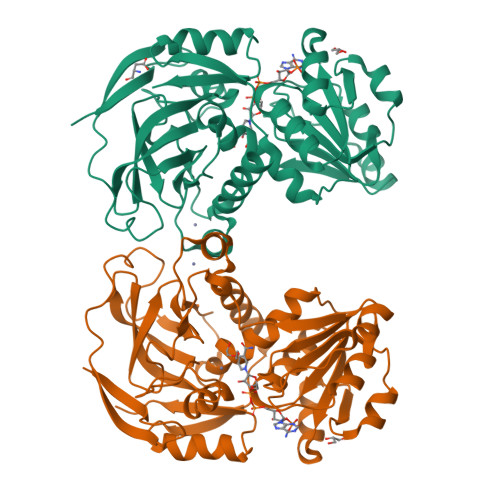
Structural Basis for Broad Substrate Selectivity of Alcohol Dehydrogenase YjgB from Escherichia coli .
Nguyen, G.T., Kim, Y.G., Ahn, J.W., Chang, J.H.(2020) Molecules 25
- PubMed: 32455802
- DOI: https://doi.org/10.3390/molecules25102404
- Primary Citation of Related Structures:
7BU2, 7BU3 - PubMed Abstract:
In metabolic engineering and synthetic biology fields, there have been efforts to produce variable bioalcohol fuels, such as isobutanol and 2-phenylethanol, in order to meet industrial demands. YjgB is an aldehyde dehydrogenase from Escherichia coli that shows nicotinamide adenine dinucleotide phosphate (NADP)-dependent broad selectivity for aldehyde derivatives with an aromatic ring or small aliphatic chain. This could contribute to the design of industrial synthetic pathways. We determined the crystal structures of YjgB for both its apo-form and NADP-complexed form at resolutions of 1.55 and 2.00 Å, respectively, in order to understand the mechanism of broad substrate selectivity. The hydrophobic pocket of the active site and the nicotinamide ring of NADP(H) are both involved in conferring its broad specificity toward aldehyde substrates. In addition, based on docking-simulation data, we inferred that π-π stacking between substrates and aromatic side chains might play a crucial role in recognizing substrates. Our structural analysis of YjgB might provide insights into establishing frameworks to understand its broad substrate specificity and develop engineered enzymes for industrial biofuel synthesis.
Organizational Affiliation:
Department of Biology Education, Kyungpook National University, 80 Daehak-ro, Buk-gu, Daegu 41566, Korea.