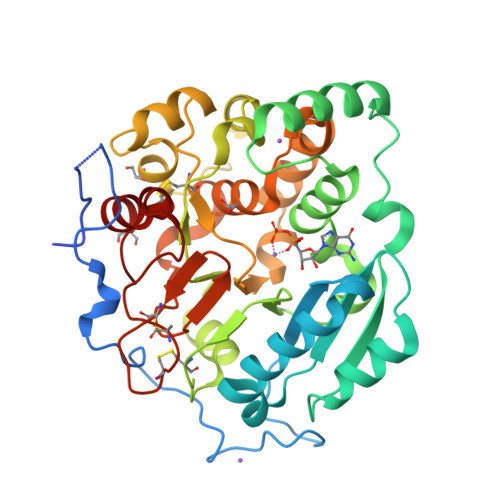
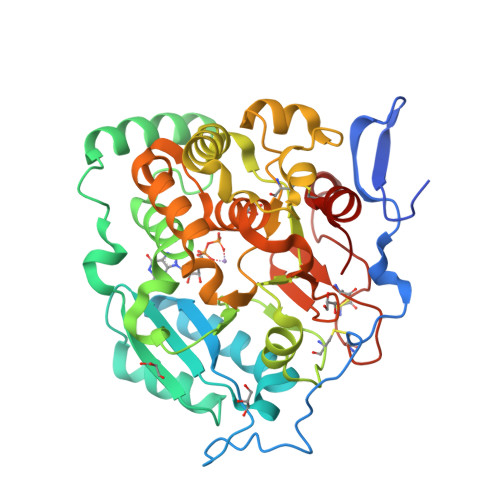
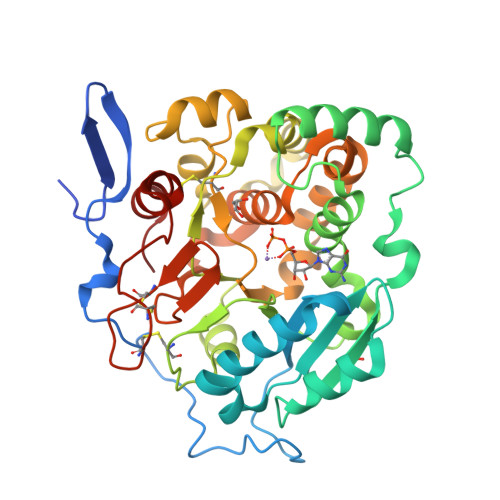
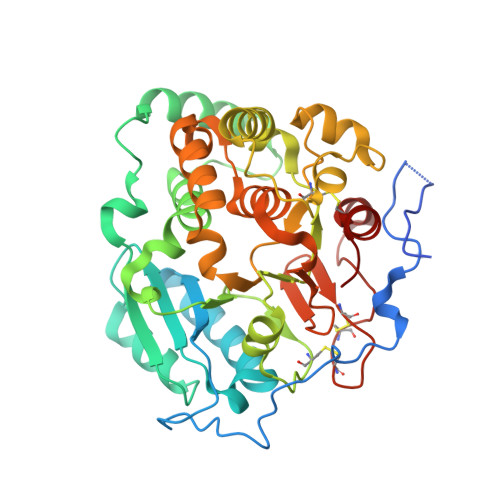
Structural basis for the core-mannan biosynthesis of cell wall fungal-type galactomannan in Aspergillus fumigatus .
Hira, D., Onoue, T., Oka, T.(2020) J Biological Chem 295: 15407-15417
- PubMed: 32873705
- DOI: https://doi.org/10.1074/jbc.RA120.013742
- Primary Citation of Related Structures:
7BOO, 7BOP - PubMed Abstract:
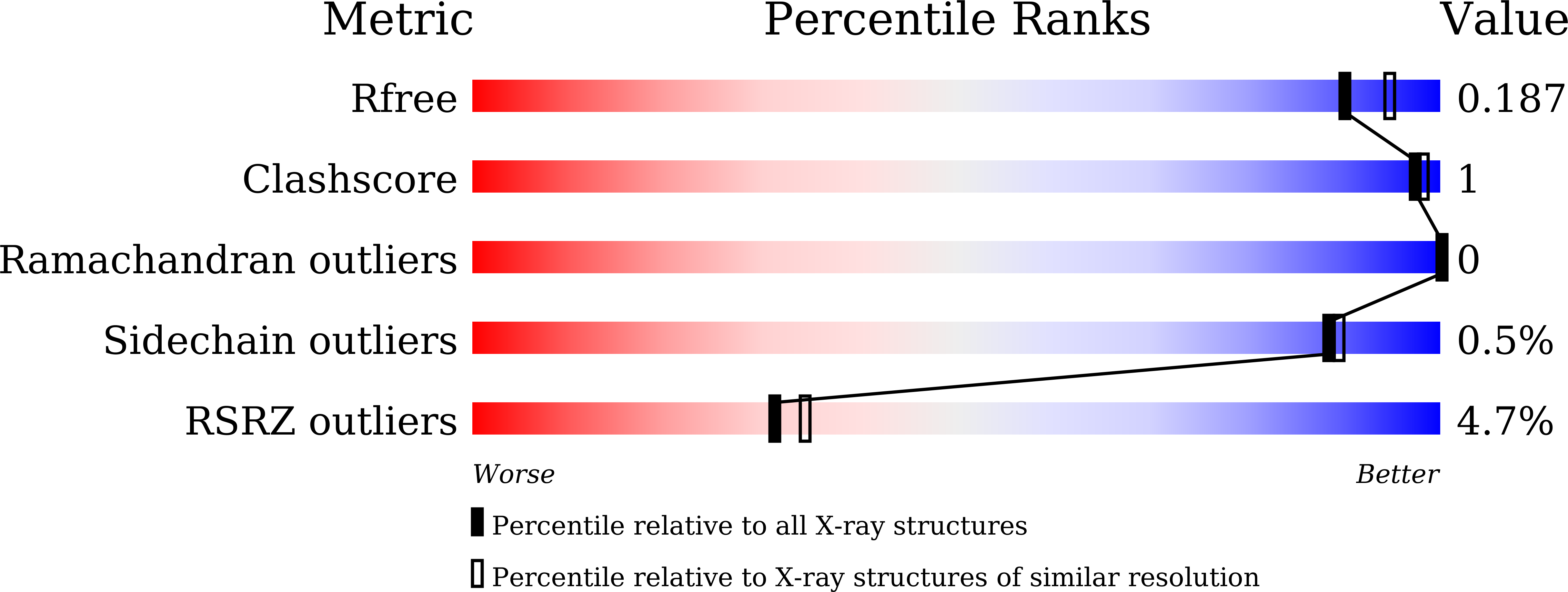
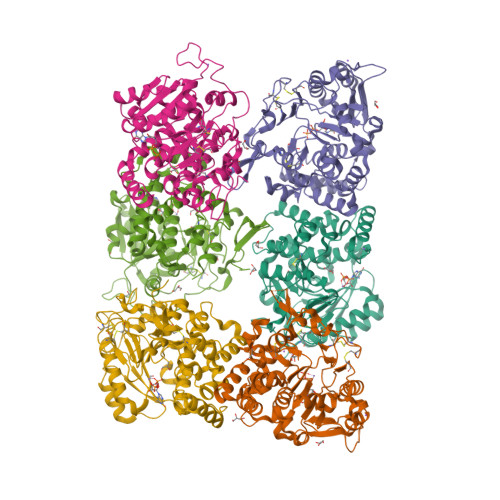
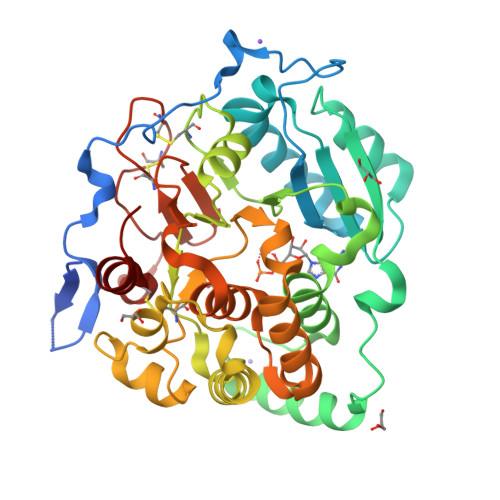
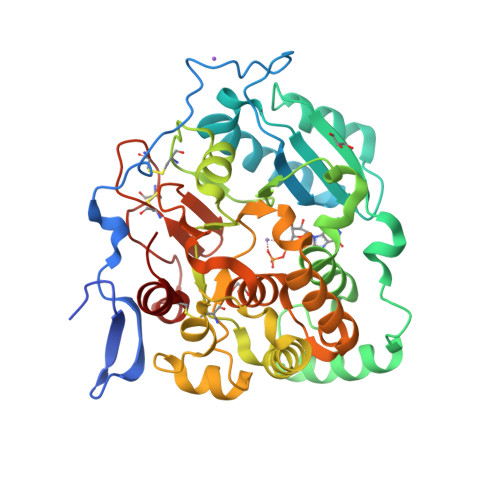
Fungal cell walls and their biosynthetic enzymes are potential targets for novel antifungal agents. Recently, two mannosyltransferases, namely core-mannan synthases A (CmsA/Ktr4) and B (CmsB/Ktr7), were found to play roles in the core-mannan biosynthesis of fungal-type galactomannan. CmsA/Ktr4 is an α-(1→2)-mannosyltransferase responsible for α-(1→2)-mannan biosynthesis in fungal-type galactomannan, which covers the cell surface of Aspergillus fumigatus Strains with disrupted cmsA / ktr4 have been shown to exhibit strongly suppressed hyphal elongation and conidiation alongside reduced virulence in a mouse model of invasive aspergillosis, indicating that CmsA/Ktr4 is a potential novel antifungal candidate. In this study we present the 3D structures of the soluble catalytic domain of CmsA/Ktr4, as determined by X-ray crystallography at a resolution of 1.95 Å, as well as the enzyme and Mn 2+ /GDP complex to 1.90 Å resolution. The CmsA/Ktr4 protein not only contains a highly conserved binding pocket for the donor substrate, GDP-mannose, but also has a unique broad cleft structure formed by its N- and C-terminal regions and is expected to recognize the acceptor substrate, a mannan chain. Based on these crystal structures, we also present a 3D structural model of the enzyme-substrate complex generated using docking and molecular dynamics simulations with α-Man-(1→6)-α-Man-(1→2)-α-Man-OMe as the model structure for the acceptor substrate. This predicted enzyme-substrate complex structure is also supported by findings from single amino acid substitution CmsA/Ktr4 mutants expressed in Δ cmsA/ktr4 A. fumigatus cells. Taken together, these results provide basic information for developing specific α-mannan biosynthesis inhibitors for use as pharmaceuticals and/or pesticides.
Organizational Affiliation:
Department of Applied Life Science, Faculty of Biotechnology and Life Science, Sojo University, Kumamoto, Kumamoto, Japan. Electronic address: hira@life.sojo-u.ac.jp.