Novel Inhibitors of Nicotinamide- N -Methyltransferase for the Treatment of Metabolic Disorders.
Kannt, A., Rajagopal, S., Hallur, M.S., Swamy, I., Kristam, R., Dhakshinamoorthy, S., Czech, J., Zech, G., Schreuder, H., Ruf, S.(2021) Molecules 26
- PubMed: 33668468
- DOI: https://doi.org/10.3390/molecules26040991
- Primary Citation of Related Structures:
7BKG, 7BLE, 7NBJ, 7NBM, 7NBQ - PubMed Abstract:
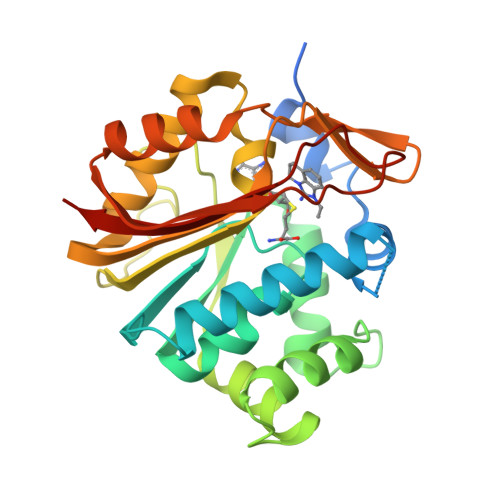
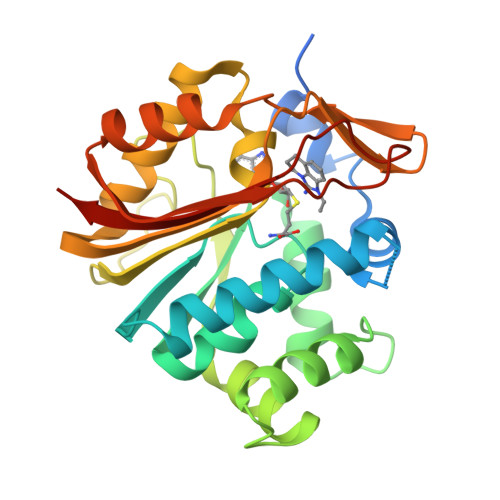
Nicotinamide- N -methyltransferase (NNMT) is a cytosolic enzyme catalyzing the transfer of a methyl group from S -adenosyl-methionine (SAM) to nicotinamide (Nam). It is expressed in many tissues including the liver, adipose tissue, and skeletal muscle. Its expression in several cancer cell lines has been widely discussed in the literature, and recent work established a link between NNMT expression and metabolic diseases. Here we describe our approach to identify potent small molecule inhibitors of NNMT featuring different binding modes as elucidated by X-ray crystallographic studies.
Organizational Affiliation:
Fraunhofer Institute for Translational Medicine and Pharmacology-ITMP, Theodor-Stern-Kai 7, 60596 Frankfurt am Main, Germany.