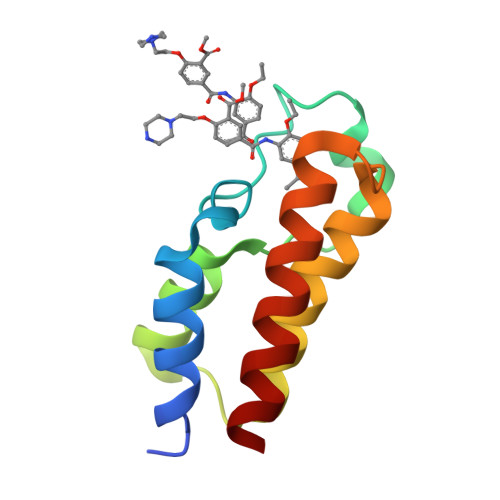
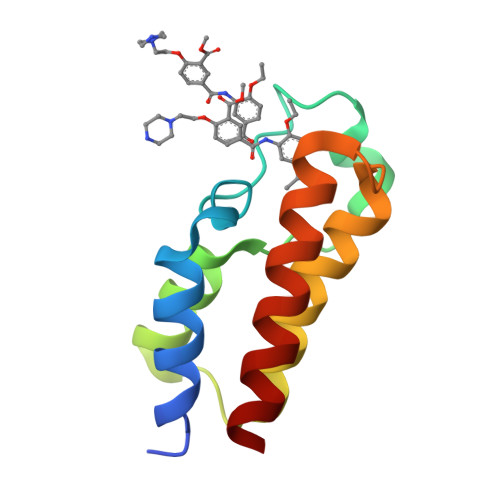
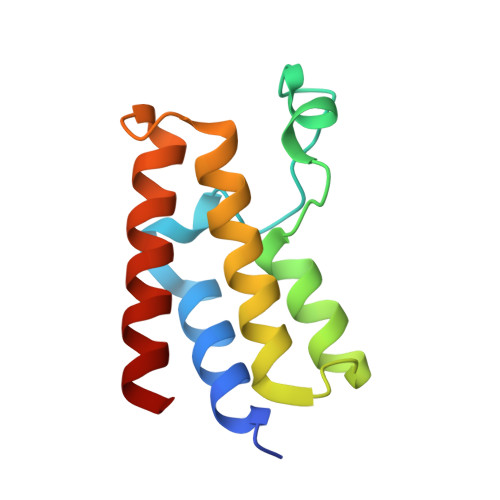
Reevaluation of bromodomain ligands targeting BAZ2A.
Cazzanelli, G., Vedove, A.D., Parolin, E., D'Agostino, V.G., Unzue, A., Nevado, C., Caflisch, A., Lolli, G.(2023) Protein Sci 32: e4752-e4752
- PubMed: 37574751
- DOI: https://doi.org/10.1002/pro.4752
- Primary Citation of Related Structures:
7BL8, 7BL9, 7BLA, 7BLB, 7BLC, 7BLD - PubMed Abstract:
BAZ2A promotes migration and invasion in prostate cancer. Two chemical probes, the specific BAZ2-ICR, and the BAZ2/BRD9 cross-reactive GSK2801, interfere with the recognition of acetylated lysines in histones by the bromodomains of BAZ2A and of its BAZ2B paralog. The two chemical probes were tested in prostate cancer cell lines with opposite androgen susceptibility. BAZ2-ICR and GSK2801 showed different cellular efficacies in accordance with their unequal selectivity profiles. Concurrent inhibition of BAZ2 and BRD9 did not reproduce the effects observed with GSK2801, indicating possible off-targets for this chemical probe. On the other hand, the single BAZ2 inhibition by BAZ2-ICR did not phenocopy genetic ablation, demonstrating that bromodomain interference is not sufficient to strongly affect BAZ2A functionality and suggesting a PROTAC-based chemical ablation as an alternative optimization strategy and a possible therapeutic approach. In this context, we also present the crystallographic structures of BAZ2A in complex with the above chemical probes. Binding poses of TP-238 and GSK4027, chemical probes for the bromodomain subfamily I, and two ligands of the CBP/EP300 bromodomains identify additional headgroups for the development of BAZ2A ligands.
Organizational Affiliation:
Department of Cellular, Computational and Integrative Biology-CIBIO, University of Trento, Trento, Italy.