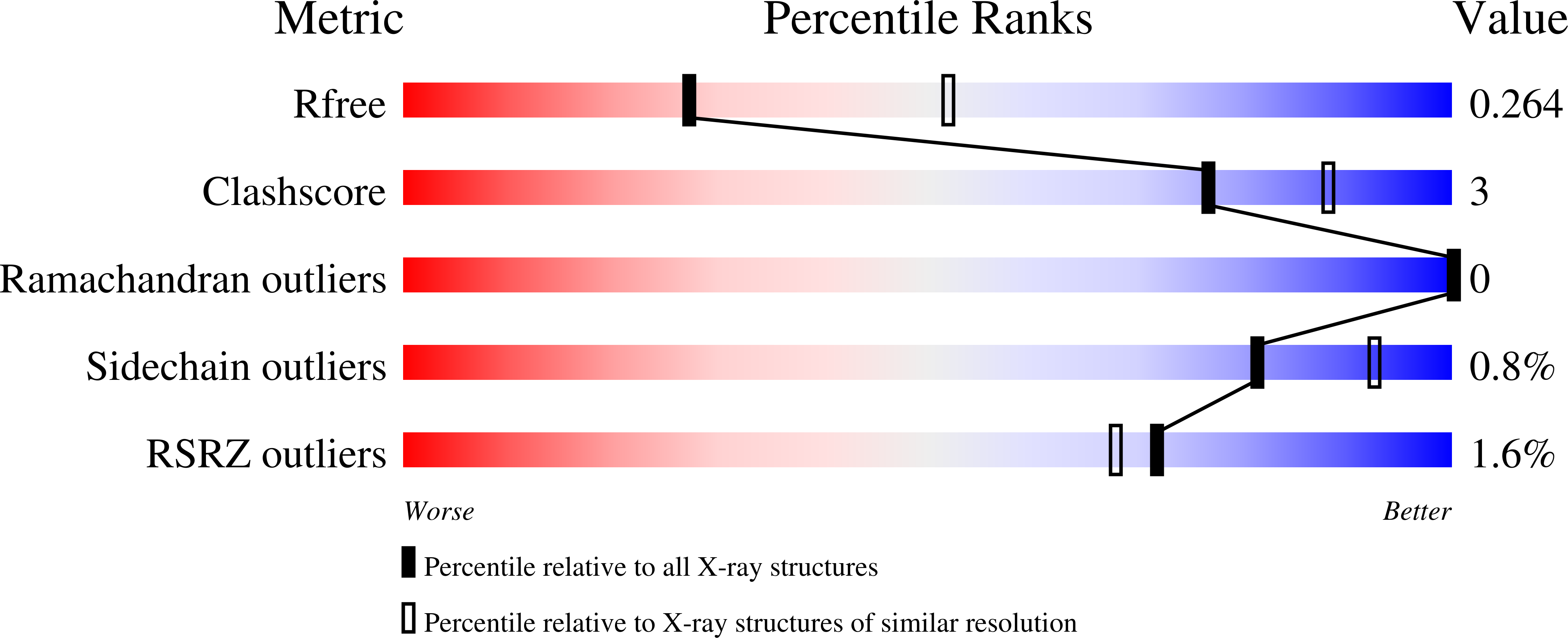
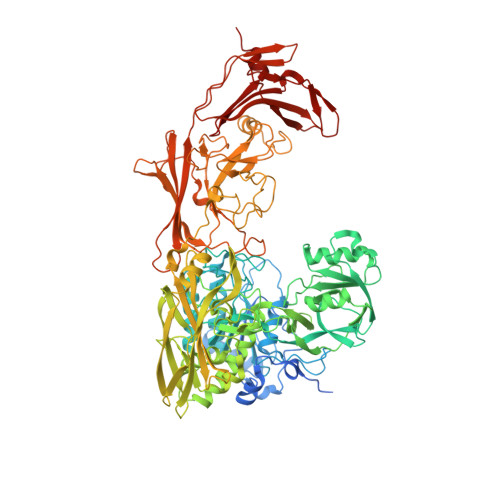
Enzyme kinetic and binding studies identify determinants of specificity for the immunomodulatory enzyme ScpA, a C5a inactivating bacterial protease.
Tecza, M., Kagawa, T.F., Jain, M., Cooney, J.C.(2021) Comput Struct Biotechnol J 19: 2356-2365
- PubMed: 33897974
- DOI: https://doi.org/10.1016/j.csbj.2021.04.024
- Primary Citation of Related Structures:
7BJ3 - PubMed Abstract:
The Streptococcal C5a peptidase (ScpA) specifically inactivates the human complement factor hC5a, a potent anaphylatoxin recently identified as a therapeutic target for treatment of COVID-19 infections. Biologics used to modulate hC5a are predominantly monoclonal antibodies. Here we present data to support an alternative therapeutic approach based on the specific inactivation of hC5a by ScpA in studies using recombinant hC5a (rhC5a). Initial characterization of ScpA confirmed activity in human serum and against rhC5a desArg (rhC5a dR ), the predominant hC5a form in blood. A new FRET based enzyme assay showed that ScpA cleaved rhC5a at near physiological concentrations ( K m 185 nM). Surface Plasmon Resonance (SPR) and Isothermal Titration Calorimetry (ITC) studies established a high affinity ScpA-rhC5a interaction ( K D 34 nM,
D ITC
Organizational Affiliation:
Department of Chemical Sciences, University of Limerick, Limerick, Ireland.