Design, Synthesis, and Biological Evaluation of Imidazopyridines as PD-1/PD-L1 Antagonists.
Butera, R., Wazynska, M., Magiera-Mularz, K., Plewka, J., Musielak, B., Surmiak, E., Sala, D., Kitel, R., de Bruyn, M., Nijman, H.W., Elsinga, P.H., Holak, T.A., Domling, A.(2021) ACS Med Chem Lett 12: 768-773
- PubMed: 34055224
- DOI: https://doi.org/10.1021/acsmedchemlett.1c00033
- Primary Citation of Related Structures:
7BEA - PubMed Abstract:
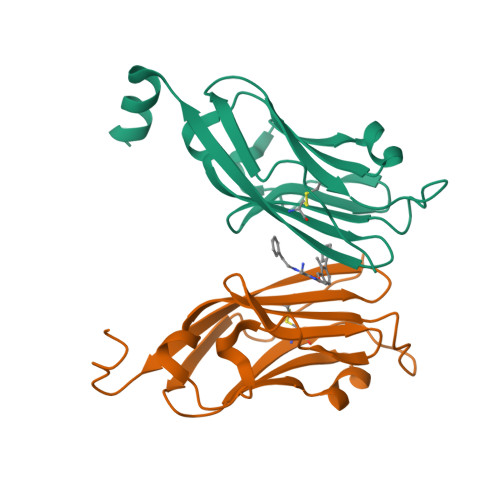
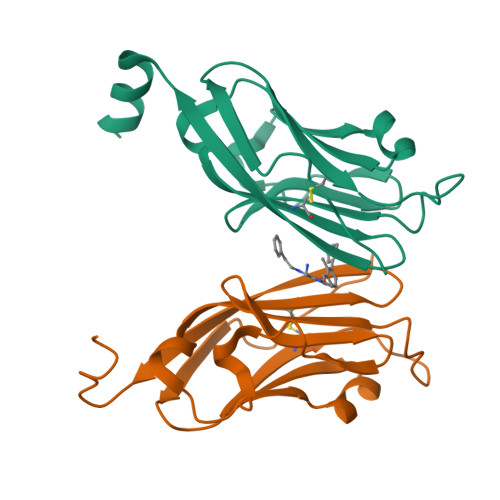
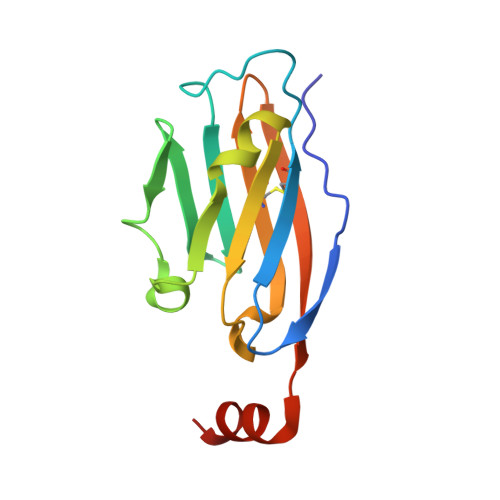
The PD-1/PD-L1 axis has proven to be a highly efficacious target for cancer immune checkpoint therapy with several approved antibodies. Also, small molecules based on a biphenyl core can antagonize PD-1/PD-L1, leading to the in vitro formation of PD-L1 dimers. However, their development remains challenging, as we do not yet fully understand their mode of action. In this work, we designed a new scaffold based on our previously solved high-resolution structures of low-molecular-weight inhibitors bound to PD-L1. A small compound library was synthesized using the Groebke-Blackburn-Bienaymé multicomponent reaction (GBB-3CR), resulting in the structure-activity relationship of imidazo[1,2- a ]pyridine-based inhibitors. These inhibitors were tested for their biological activity using various biophysical assays giving potent candidates with low-micromolar PD-L1 affinities. An obtained PD-L1 cocrystal structure reveals the binding to PD-L1. Our results open the door to an interesting bioactive scaffold that could lead to a new class of PD-L1 antagonists.
Organizational Affiliation:
Department of Drug Design, University of Groningen, A. Deusinglaan 1, 9713 AV Groningen, The Netherlands.