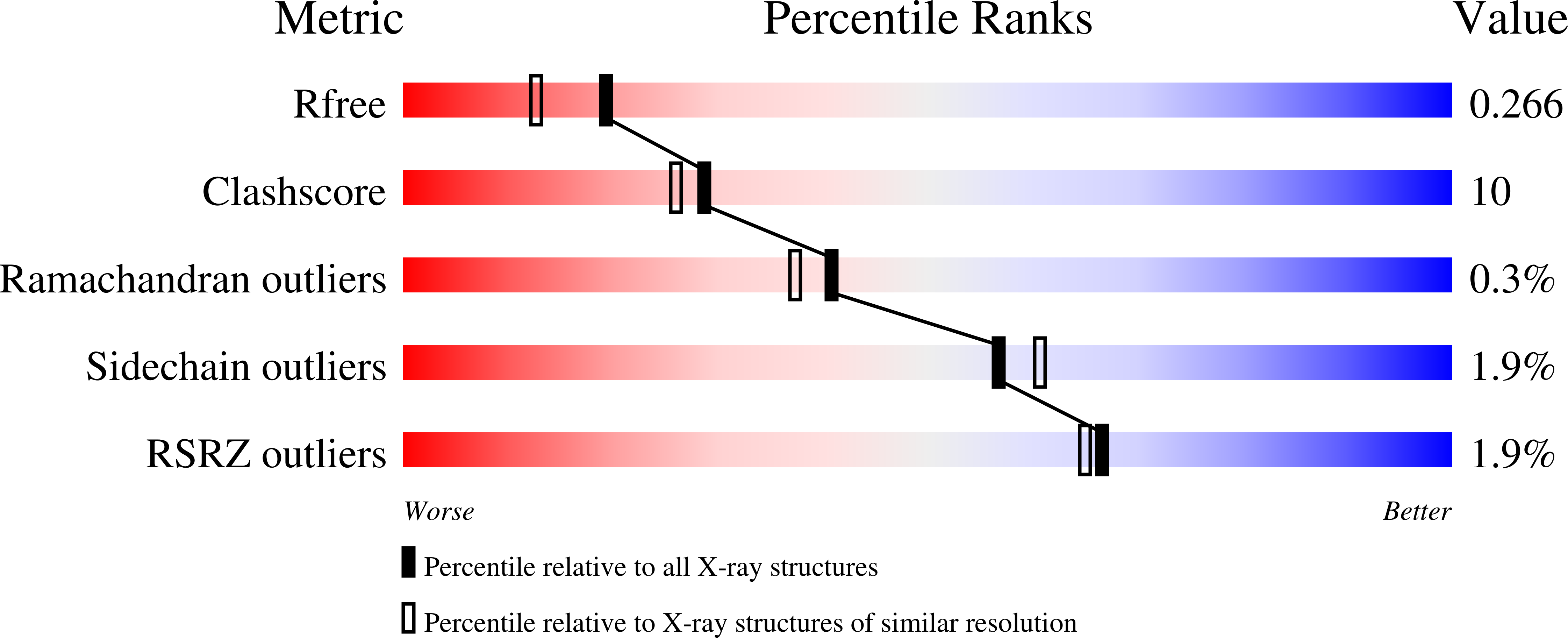
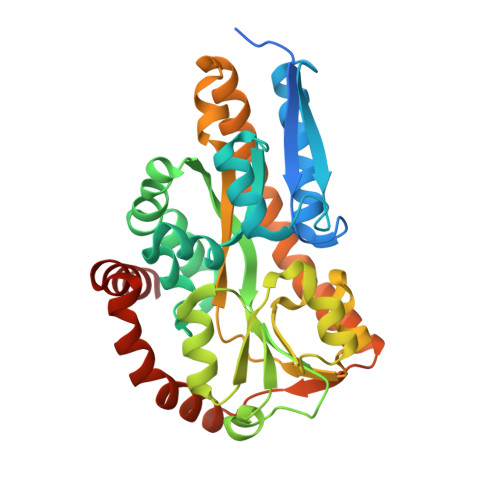
Crystal structure of the sugar acid-binding protein CxaP from a TRAP transporter in Advenella mimigardefordensis strain DPN7 T .
Schafer, L., Meinert-Berning, C., Kobus, S., Hoppner, A., Smits, S.H.J., Steinbuchel, A.(2021) FEBS J 288: 4905-4917
- PubMed: 33630388
- DOI: https://doi.org/10.1111/febs.15789
- Primary Citation of Related Structures:
7BBR, 7BCN, 7BCO, 7BCP, 7BCR - PubMed Abstract:
Recently, CxaP, a sugar acid substrate binding protein (SBP) from Advenella mimigardefordensis strain DPN7 T , was identified as part of a novel sugar uptake strategy. In the present study, the protein was successfully crystallized. Although several SBP structures of tripartite ATP-independent periplasmic transporters have already been solved, this is the first structure of an SBP accepting multiple sugar acid ligands. Protein crystals were obtained with bound d-xylonic acid, d-fuconic acid d-galactonic and d-gluconic acid, respectively. The protein shows the typical structure of an SBP of a tripartite ATP-independent periplasmic transporter consisting of two domains linked by a hinge and spanned by a long α-helix. By analysis of the structure, the substrate binding site of the protein was identified. The carboxylic group of the sugar acids interacts with Arg175, whereas the coordination of the hydroxylic groups at positions C2 and C3 is most probably realized by Arg154 and Asn151. Furthermore, it was observed that 2-keto-3-deoxy-d-gluconic acid is bound in protein crystals that were crystallized without the addition of any ligand, indicating that this molecule is prebound to the protein and is displaced by the other ligands if they are available. DATABASE: Structural data of CxaP complexes are available in the worldwide Protein Data Bank (https://www.rcsb.org) under the accession codes 7BBR (2-keto-3-deoxy-d-gluconic acid), 7BCR (d-galactonic acid), 7BCN (d-xylonic acid), 7BCO (d-fuconic acid) and 7BCP (d-gluconic acid).
Organizational Affiliation:
Institute of Molecular Microbiology and Biotechnology, Westfälische Wilhelms University Münster, Münster, Germany.