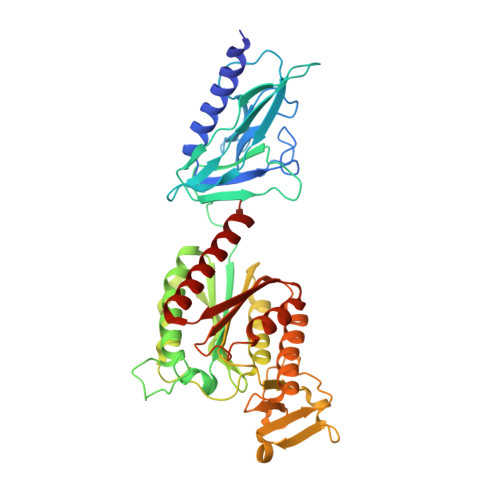
PilB from Streptococcus sanguinis is a bimodular type IV pilin with a direct role in adhesion.
Raynaud, C., Sheppard, D., Berry, J.L., Gurung, I., Pelicic, V.(2021) Proc Natl Acad Sci U S A 118
- PubMed: 34031252
- DOI: https://doi.org/10.1073/pnas.2102092118
- Primary Citation of Related Structures:
7B7P, 7BA2 - PubMed Abstract:
Type IV pili (T4P) are functionally versatile filamentous nanomachines, nearly ubiquitous in prokaryotes. They are predominantly polymers of one major pilin but also contain minor pilins whose functions are often poorly defined and likely to be diverse. Here, we show that the minor pilin PilB from the T4P of Streptococcus sanguinis displays an unusual bimodular three-dimensional structure with a bulky von Willebrand factor A-like (vWA) module "grafted" onto a small pilin module via a short loop. Structural modeling suggests that PilB is only compatible with a localization at the tip of T4P. By performing a detailed functional analysis, we found that 1) the vWA module contains a canonical metal ion-dependent adhesion site, preferentially binding Mg 2+ and Mn 2+ , 2) abolishing metal binding has no impact on the structure of PilB or piliation, 3) metal binding is important for S. sanguinis T4P-mediated twitching motility and adhesion to eukaryotic cells, and 4) the vWA module shows an intrinsic binding ability to several host proteins. These findings reveal an elegant yet simple evolutionary tinkering strategy to increase T4P functional versatility by grafting a functional module onto a pilin for presentation by the filaments. This strategy appears to have been extensively used by bacteria, in which modular pilins are widespread and exhibit an astonishing variety of architectures.
Organizational Affiliation:
Medical Research Council Centre for Molecular Bacteriology and Infection, Imperial College London, London SW7 2AZ, United Kingdom.