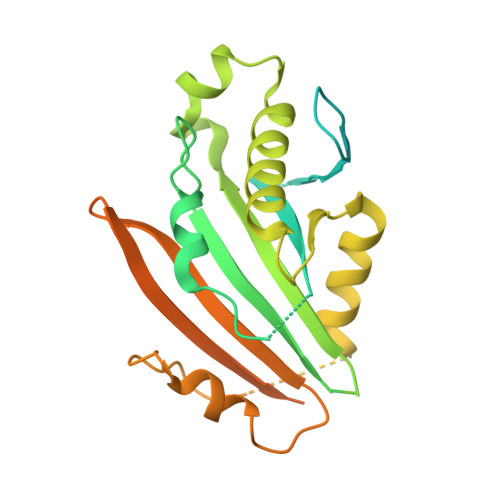
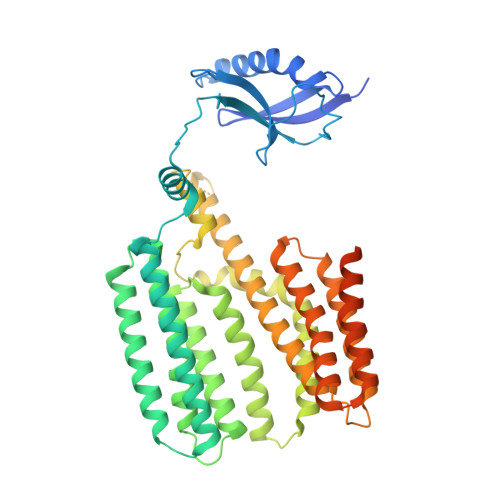
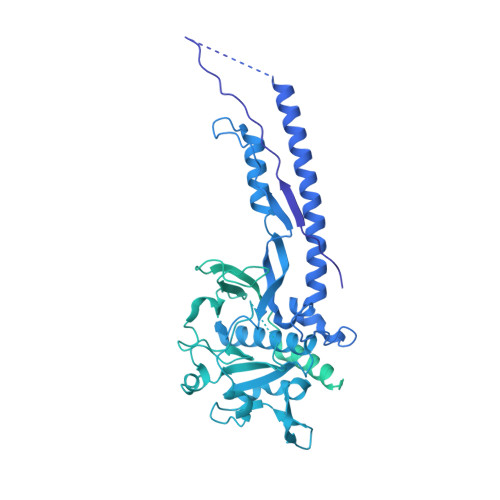
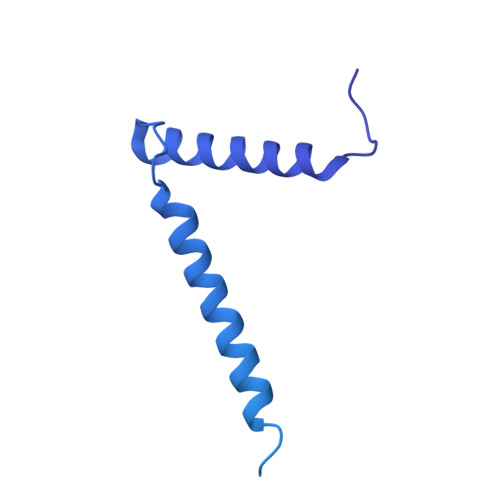
Structure of the mycobacterial ESX-5 type VII secretion system pore complex.
Beckham, K.S.H., Ritter, C., Chojnowski, G., Ziemianowicz, D.S., Mullapudi, E., Rettel, M., Savitski, M.M., Mortensen, S.A., Kosinski, J., Wilmanns, M.(2021) Sci Adv 7
- PubMed: 34172453
- DOI: https://doi.org/10.1126/sciadv.abg9923
- Primary Citation of Related Structures:
7B7J, 7B9F, 7B9S - PubMed Abstract:
The ESX-5 type VII secretion system is a membrane-spanning protein complex key to the virulence of mycobacterial pathogens. However, the overall architecture of the fully assembled translocation machinery and the composition of the central secretion pore have remained unknown. Here, we present the high-resolution structure of the 2.1-megadalton ESX-5 core complex. Our structure captured a dynamic, secretion-competent conformation of the pore within a well-defined transmembrane section, sandwiched between two flexible protein layers at the cytosolic entrance and the periplasmic exit. We propose that this flexibility endows the ESX-5 machinery with large conformational plasticity required to accommodate targeted protein secretion. Compared to known secretion systems, a highly dynamic state of the pore may represent a fundamental principle of bacterial secretion machineries.
Organizational Affiliation:
European Molecular Biology Laboratory, Hamburg Unit, Notkestrasse 85, 22607 Hamburg, Germany.