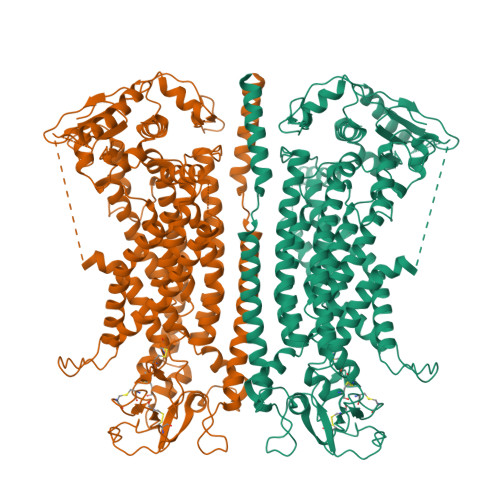
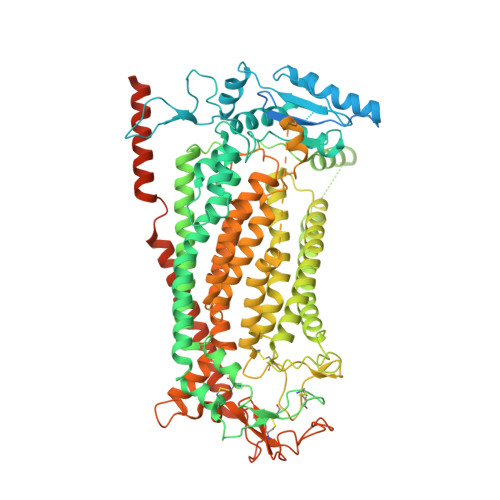
Gating the pore of the calcium-activated chloride channel TMEM16A.
Lam, A.K.M., Rheinberger, J., Paulino, C., Dutzler, R.(2021) Nat Commun 12: 785-785
- PubMed: 33542223
- DOI: https://doi.org/10.1038/s41467-020-20787-9
- Primary Citation of Related Structures:
7B5C, 7B5D, 7B5E - PubMed Abstract:
The binding of cytoplasmic Ca 2+ to the anion-selective channel TMEM16A triggers a conformational change around its binding site that is coupled to the release of a gate at the constricted neck of an hourglass-shaped pore. By combining mutagenesis, electrophysiology, and cryo-electron microscopy, we identified three hydrophobic residues at the intracellular entrance of the neck as constituents of this gate. Mutation of each of these residues increases the potency of Ca 2+ and results in pronounced basal activity. The structure of an activating mutant shows a conformational change of an α-helix that contributes to Ca 2+ binding as a likely cause for the basal activity. Although not in physical contact, the three residues are functionally coupled to collectively contribute to the stabilization of the gate in the closed conformation of the pore, thus explaining the low open probability of the channel in the absence of Ca 2+ .
Organizational Affiliation:
Department of Biochemistry, University of Zurich, Winterthurerstrasse 190, CH-8057, Zurich, Switzerland. a.lam@bioc.uzh.ch.