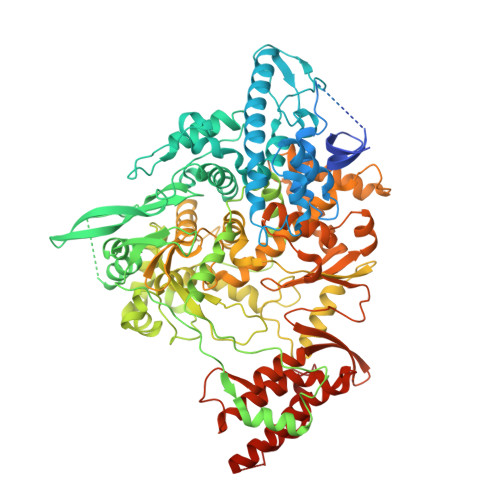
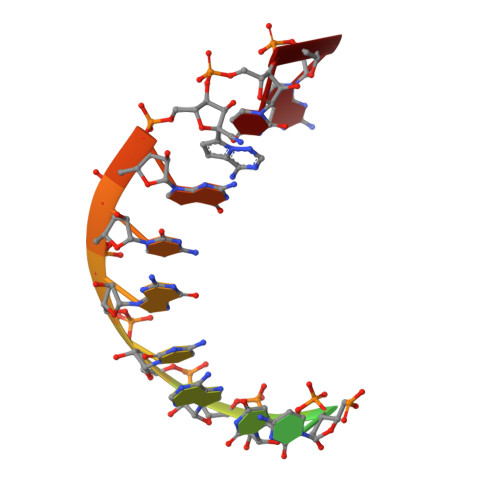
Mechanism of SARS-CoV-2 polymerase stalling by remdesivir.
Kokic, G., Hillen, H.S., Tegunov, D., Dienemann, C., Seitz, F., Schmitzova, J., Farnung, L., Siewert, A., Hobartner, C., Cramer, P.(2021) Nat Commun 12: 279-279
- PubMed: 33436624
- DOI: https://doi.org/10.1038/s41467-020-20542-0
- Primary Citation of Related Structures:
7B3B, 7B3C, 7B3D - PubMed Abstract:
Remdesivir is the only FDA-approved drug for the treatment of COVID-19 patients. The active form of remdesivir acts as a nucleoside analog and inhibits the RNA-dependent RNA polymerase (RdRp) of coronaviruses including SARS-CoV-2. Remdesivir is incorporated by the RdRp into the growing RNA product and allows for addition of three more nucleotides before RNA synthesis stalls. Here we use synthetic RNA chemistry, biochemistry and cryo-electron microscopy to establish the molecular mechanism of remdesivir-induced RdRp stalling. We show that addition of the fourth nucleotide following remdesivir incorporation into the RNA product is impaired by a barrier to further RNA translocation. This translocation barrier causes retention of the RNA 3'-nucleotide in the substrate-binding site of the RdRp and interferes with entry of the next nucleoside triphosphate, thereby stalling RdRp. In the structure of the remdesivir-stalled state, the 3'-nucleotide of the RNA product is matched and located with the template base in the active center, and this may impair proofreading by the viral 3'-exonuclease. These mechanistic insights should facilitate the quest for improved antivirals that target coronavirus replication.
- Max Planck Institute for Biophysical Chemistry, Department of Molecular Biology, Am Fassberg 11, Göttingen, 37077, Germany.
Organizational Affiliation: