Molecular mechanism of Mad1 kinetochore targeting by phosphorylated Bub1.
Fischer, E.S., Yu, C.W.H., Bellini, D., McLaughlin, S.H., Orr, C.M., Wagner, A., Freund, S.M.V., Barford, D.(2021) EMBO Rep 22: e52242-e52242
- PubMed: 34013668
- DOI: https://doi.org/10.15252/embr.202052242
- Primary Citation of Related Structures:
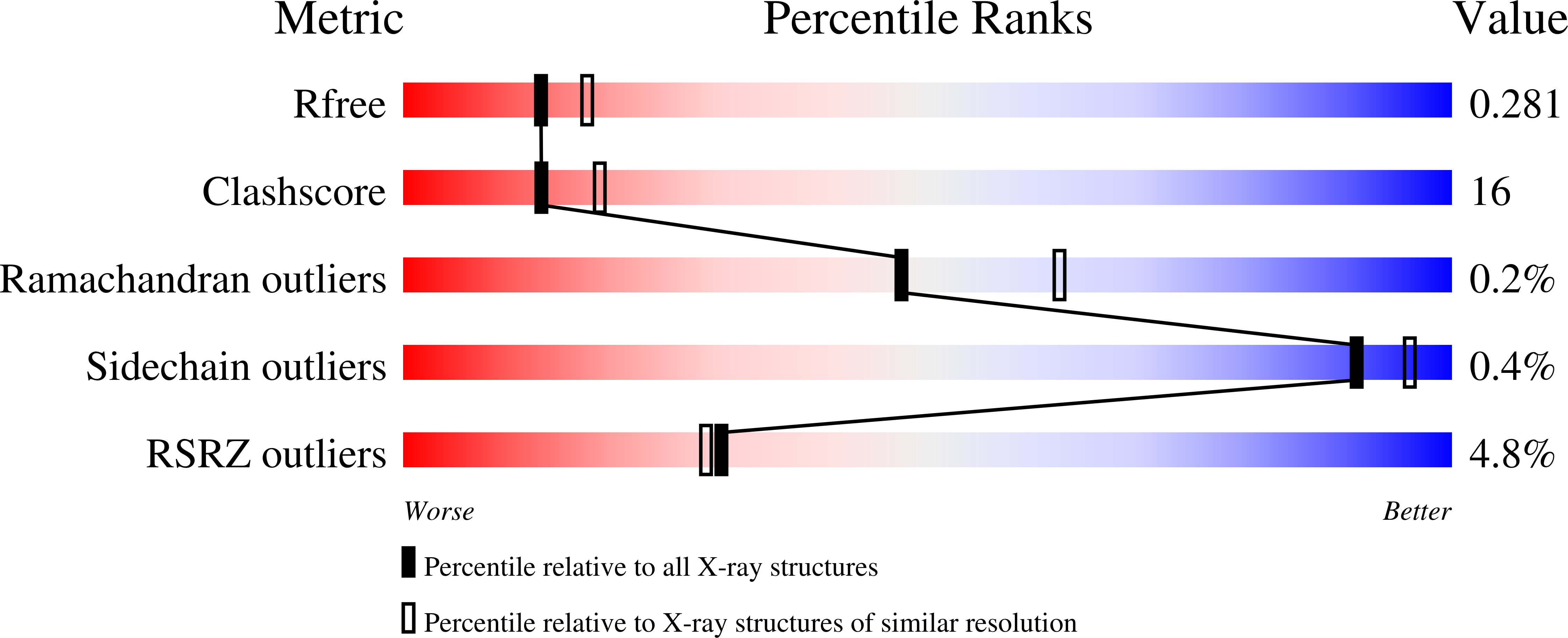
7B1F, 7B1H, 7B1J - PubMed Abstract:
During metaphase, in response to improper kinetochore-microtubule attachments, the spindle assembly checkpoint (SAC) activates the mitotic checkpoint complex (MCC), an inhibitor of the anaphase-promoting complex/cyclosome (APC/C). This process is orchestrated by the kinase Mps1, which initiates the assembly of the MCC onto kinetochores through a sequential phosphorylation-dependent signalling cascade. The Mad1-Mad2 complex, which is required to catalyse MCC formation, is targeted to kinetochores through a direct interaction with the phosphorylated conserved domain 1 (CD1) of Bub1. Here, we present the crystal structure of the C-terminal domain of Mad1 (Mad1 CTD ) bound to two phosphorylated Bub1 CD1 peptides at 1.75 Å resolution. This interaction is mediated by phosphorylated Bub1 Thr461, which not only directly interacts with Arg617 of the Mad1 RLK (Arg-Leu-Lys) motif, but also directly acts as an N-terminal cap to the CD1 α-helix dipole. Surprisingly, only one Bub1 CD1 peptide binds to the Mad1 homodimer in solution. We suggest that this stoichiometry is due to inherent asymmetry in the coiled-coil of Mad1 CTD and has implications for how the Mad1-Bub1 complex at kinetochores promotes efficient MCC assembly.
Organizational Affiliation:
MRC Laboratory of Molecular Biology, Cambridge, UK.