Structure-Based Optimization and Discovery of M3258, a Specific Inhibitor of the Immunoproteasome Subunit LMP7 ( beta 5i).
Klein, M., Busch, M., Friese-Hamim, M., Crosignani, S., Fuchss, T., Musil, D., Rohdich, F., Sanderson, M.P., Seenisamy, J., Walter-Bausch, G., Zanelli, U., Hewitt, P., Esdar, C., Schadt, O.(2021) J Med Chem 64: 10230-10245
- PubMed: 34228444
- DOI: https://doi.org/10.1021/acs.jmedchem.1c00604
- Primary Citation of Related Structures:
7B12 - PubMed Abstract:
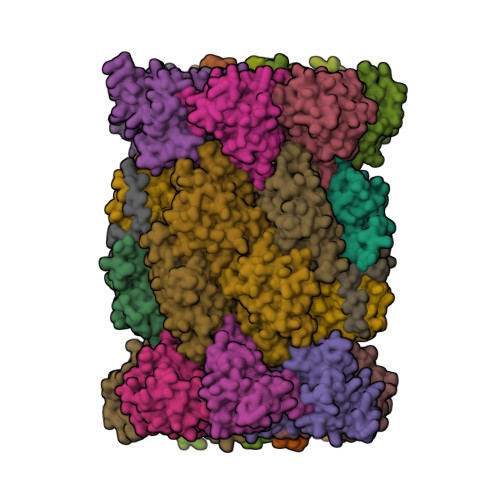
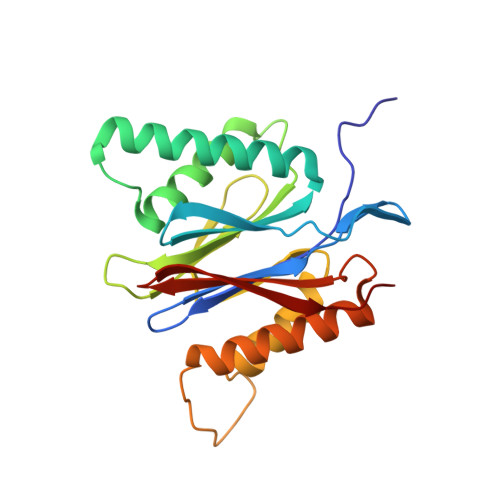
Proteasomes are broadly expressed key components of the ubiquitin-dependent protein degradation pathway containing catalytically active subunits (β1, β2, and β5). LMP7 (β5i) is a subunit of the immunoproteasome, an inducible isoform that is predominantly expressed in hematopoietic cells. Clinically effective pan-proteasome inhibitors for the treatment of multiple myeloma (MM) nonselectively target LMP7 and other subunits of the constitutive proteasome and immunoproteasome with comparable potency, which can limit the therapeutic applicability of these drugs. Here, we describe the discovery and structure-based hit optimization of novel amido boronic acids, which selectively inhibit LMP7 while sparing all other subunits. The exploitation of structural differences between the proteasome subunits culminated in the identification of the highly potent, exquisitely selective, and orally available LMP7 inhibitor 50 (M3258). Based on the strong antitumor activity observed with M3258 in MM models and a favorable preclinical data package, a phase I clinical trial was initiated in relapsed/refractory MM patients.
Organizational Affiliation:
Merck KGaA, Frankfurter Str. 250, Darmstadt 64293, Germany.