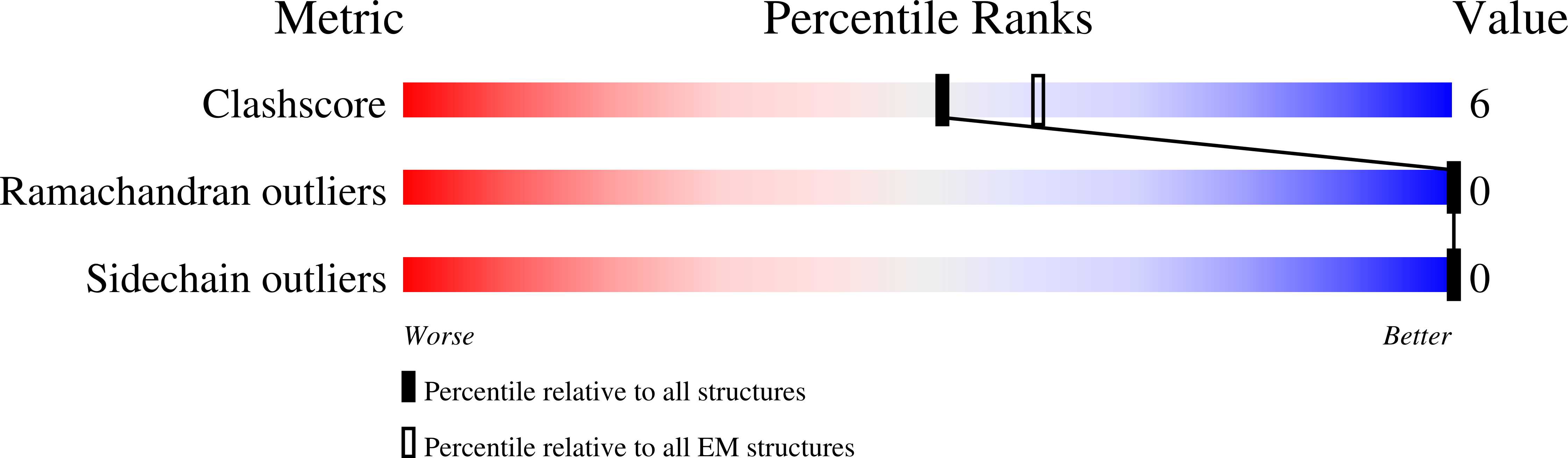Structural basis for substrate specificity of heteromeric transporters of neutral amino acids.
Rodriguez, C.F., Escudero-Bravo, P., Diaz, L., Bartoccioni, P., Garcia-Martin, C., Gilabert, J.G., Boskovic, J., Guallar, V., Errasti-Murugarren, E., Llorca, O., Palacin, M.(2021) Proc Natl Acad Sci U S A 118
- PubMed: 34848541
- DOI: https://doi.org/10.1073/pnas.2113573118
- Primary Citation of Related Structures:
7B00 - PubMed Abstract:
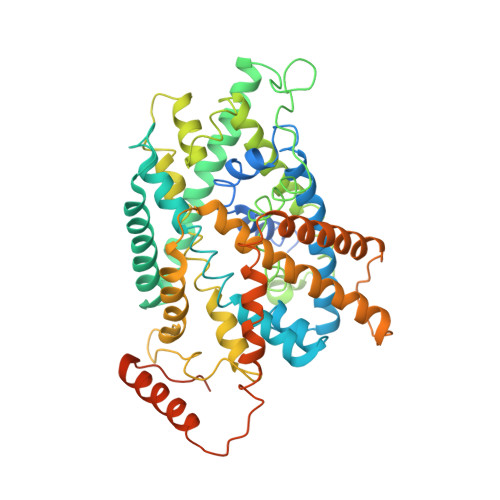
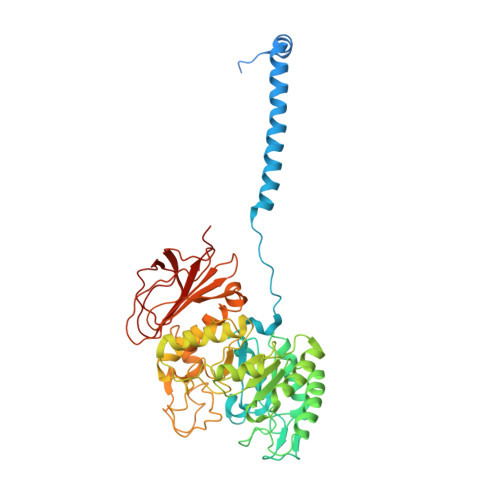
Despite having similar structures, each member of the heteromeric amino acid transporter (HAT) family shows exquisite preference for the exchange of certain amino acids. Substrate specificity determines the physiological function of each HAT and their role in human diseases. However, HAT transport preference for some amino acids over others is not yet fully understood. Using cryo-electron microscopy of apo human LAT2/CD98hc and a multidisciplinary approach, we elucidate key molecular determinants governing neutral amino acid specificity in HATs. A few residues in the substrate-binding pocket determine substrate preference. Here, we describe mutations that interconvert the substrate profiles of LAT2/CD98hc, LAT1/CD98hc, and Asc1/CD98hc. In addition, a region far from the substrate-binding pocket critically influences the conformation of the substrate-binding site and substrate preference. This region accumulates mutations that alter substrate specificity and cause hearing loss and cataracts. Here, we uncover molecular mechanisms governing substrate specificity within the HAT family of neutral amino acid transporters and provide the structural bases for mutations in LAT2/CD98hc that alter substrate specificity and that are associated with several pathologies.
Organizational Affiliation:
Structural Biology Programme, Spanish National Cancer Research Centre, E-28029 Madrid, Spain.