Selective Aster inhibitors distinguish vesicular and nonvesicular sterol transport mechanisms.
Xiao, X., Kim, Y., Romartinez-Alonso, B., Sirvydis, K., Ory, D.S., Schwabe, J.W.R., Jung, M.E., Tontonoz, P.(2021) Proc Natl Acad Sci U S A 118
- PubMed: 33376205
- DOI: https://doi.org/10.1073/pnas.2024149118
- Primary Citation of Related Structures:
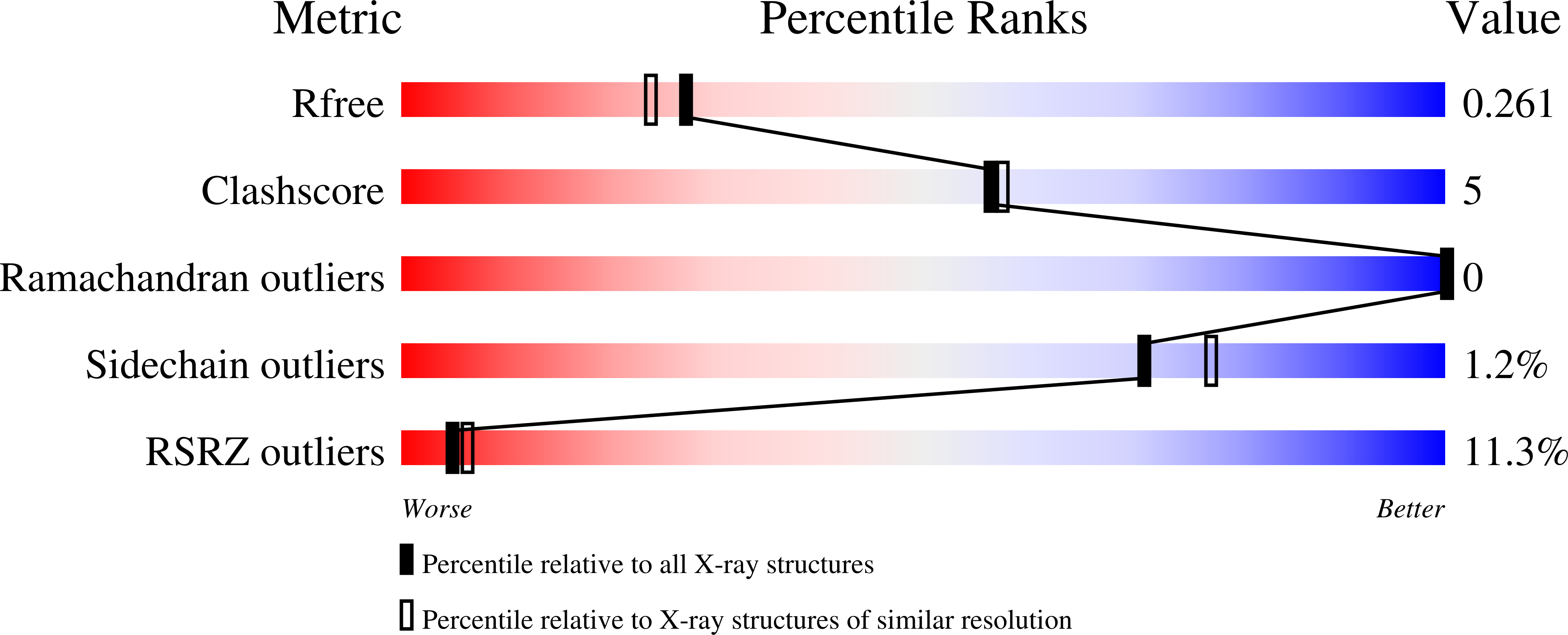
7AZN - PubMed Abstract:
The Aster proteins (encoded by the Gramd1a-c genes) contain a ligand-binding fold structurally similar to a START domain and mediate nonvesicular plasma membrane (PM) to endoplasmic reticulum (ER) cholesterol transport. In an effort to develop small molecule modulators of Asters, we identified 20α-hydroxycholesterol (HC) and U18666A as lead compounds. Unfortunately, both 20α-HC and U18666A target other sterol homeostatic proteins, limiting their utility. 20α-HC inhibits sterol regulatory element-binding protein 2 (SREBP2) processing, and U18666A is an inhibitor of the vesicular trafficking protein Niemann-Pick C1 (NPC1). To develop potent and selective Aster inhibitors, we synthesized a series of compounds by modifying 20α-HC and U18666A. Among these, AI (Aster inhibitor)-1l, which has a longer side chain than 20α-HC, selectively bound to Aster-C. The crystal structure of Aster-C in complex with AI-1l suggests that sequence and flexibility differences in the loop that gates the binding cavity may account for the ligand specificity for Aster C. We further identified the U18666A analog AI-3d as a potent inhibitor of all three Aster proteins. AI-3d blocks the ability of Asters to bind and transfer cholesterol in vitro and in cells. Importantly, AI-3d also inhibits the movement of low-density lipoprotein (LDL) cholesterol to the ER, although AI-3d does not block NPC1. This finding positions the nonvesicular Aster pathway downstream of NPC1-dependent vesicular transport in the movement of LDL cholesterol to the ER. Selective Aster inhibitors represent useful chemical tools to distinguish vesicular and nonvesicular sterol transport mechanisms in mammalian cells.
Organizational Affiliation:
Department of Pathology and Laboratory Medicine, University of California, Los Angeles, CA 90095.